Combined Educational & Scientific Session
Biophysics & Metabolism Studied with Imaging & Spectroscopy of Non-Hyperpolarized X-Nuclei
Session Topic: Other Nuclei MRI and MRS to Study Metabolism
Session Sub-Topic: Biophysics & Metabolism Studied with Imaging & Spectroscopy of Non-Hyperpolarized X-Nuclei
Combined Educational & Scientific Session
ORGANIZERS: Ronald Ouwerkerk
| Tuesday Parallel 5 Live Q&A | Tuesday, 11 August 2020, 13:45 - 14:30 UTC | Moderators: Ravinder Reddy |
Skill Level: Basic to Advanced
Session Number: C-W-01
Overview
The first part of this course covers the basics of 23Na-MR imaging, acquisition methods and applications and a brief introduction to other nuclei requiring similar acquisition strategies such as 170, 39K. The second introduces methods to study metabolic processes with 31P and deuterium MR spectroscopy.
Target Audience
Scientists and clinicians who are interested in using broadband equipped MR scanners to explore new MRI and MRS methods.
Educational Objectives
As a result of attending this course, participants should be able to:
- Give an overview of X-nuclei methods;
- Explain some of the technical challenges of X-nuclei MR;
- Describe the range of applications for biomedical research; and
- Identify new developments in X-nuclei imaging and MRS.
| Imaging Biophysics & Metabolism with Other Nuclei (23Na, 17O, 39K)
Jean-Philippe Ranjeva
In light of technical advancements supporting exploration of MR signals other than 1H, MRI of other nuclei having magnetic properties such as sodium (23Na), Oxygen (17O) or potassium (39K) is developing in parallel with the increase of the magnetic fields of clinical MR scanners.These modalities receive attention as markers of ionic homeostasis and cell viability (23Na, 39K) or provide non-invasive way to determine cerebral metabolic rate of oxygen (CMR02) consumption using 17O MRI.During this course, we will present the practical issues to conduct MRI of these particular nuclei, hypotheses and proxy to derive the biophysical parameters from these images
|
||
0472.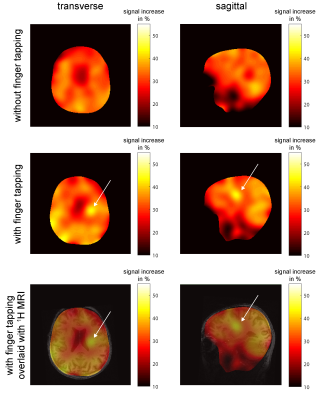 |
Mapping neuronal activity associated with finger tapping using direct measurement of 17O at 7 Tesla: proof-of-concept experiment
Tanja Platt1, Louise Ebersberger2,3, Vanessa L Franke1,4, Armin M Nagel1,5,6, Reiner Umathum1, Heinz-Peter Schlemmer2, Peter Bachert1,4, Mark E Ladd1,3,4, Andreas Korzowski1, Sebastian C Niesporek1, and Daniel Paech2
1Medical Physics in Radiology, German Cancer Research Center, Heidelberg, Germany, 2Radiology, German Cancer Research Center, Heidelberg, Germany, 3Faculty of Medicine, University of Heidelberg, Heidelberg, Germany, 4Faculty of Physics and Astronomy, University of Heidelberg, Heidelberg, Germany, 5Institute of Radiology, University Hospital Erlangen, Erlangen, Germany, 6Institute of Medical Physics, Friedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Erlangen, Germany
Dynamic 17O-MRI enables direct quantification of the cerebral metabolic rate of oxygen (CMRO2) consumption. We investigated hemispherical dependence of the method in three healthy volunteers as well as its potential for mapping neuronal activity associated with finger tapping in one healthy volunteer. Our findings were consistent with previous results, demonstrating higher CMRO2 values in gray compared to white matter. Evaluation of left/right hemispheric CMRO2 values without sensomotoric stimulation demonstrated hemispherical independence of the technique. The finger-tapping experiment demonstrated increased 17O-signal in the stimulated sensorimotor cortex and adjacent brain tissue, indicating that dynamic 17O-MRI may permit visualization of physiological neuronal activity.
|
|
0473.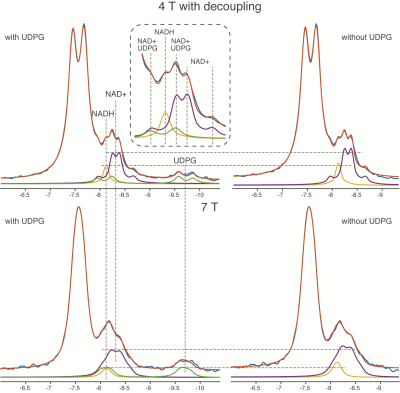 |
In vivo NAD+/NADH Measurements in the Human Brain using 31P MRS at 4 T and 7 T
Xi Chen1, Elliot Kuan2, Dost Ongur1, Wei Chen3, and Fei Du1
1McLean Hospital; Harvard Medical School, Belmont, MA, United States, 2McLean Hospital, Belmont, MA, United States, 3University of Minnesota, Minneapolis, MN, United States
Nicotinamide adenine dinucleotides play a crucial role in human health, but measuring the redox ratio (NAD+/NADH) in vivo is technically challenging and the confounding effects from UDPG remain unclear. In this study, for the first time we observed that the NAD+/NADH values decreased in 4T proton-decoupling spectra when the UDPG contribution was accounted for as well as confirming the opposite trend at 7T. Furthermore, we revealed different overlapping patterns at 4T and 7T which lead to this result. Finally, individual redox ratio measures with and without UDPG quantification are strongly correlated with one another at both 4T and 7T.
|
|
 |
0474.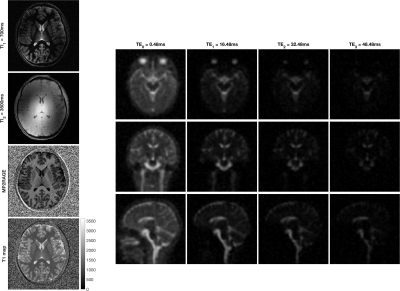 |
Multiplexed Measurement of Multiple Nuclei (M3N) for integrated multi-nuclei imaging and parametric mapping in 23Na and 1H MRI
Yasmin Blunck1,2, Daniel Staeb3, Rebecca K Glarin2,4,5, Bradford A Moffat2,4, Kieran O'Brien6, and Leigh A Johnston1,2
1Department of Biomedical Engineering, University of Melbourne, Parkville, Australia, 2Melbourne Brain Centre Imaging Unit, University of Melbourne, Parkville, Australia, 3MR Research Collaborations, Siemens Healthcare Pty Ltd, Melbourne, Australia, 4Department of Medicine & Radiology, University of Melbourne, Parkville, Australia, 5Department of Radiology, Royal Melbourne Hospital, Parkville, Australia, 6MR Research Collaborations, Siemens Healthcare Pty Ltd, Brisbane, Australia
X-nuclei imaging like sodium MRI offers complementary information to 1H-based imaging. However, its clinical translation is hampered by the significant increase in scan time required for the acquisition of an additional nuclei. Addressing this challenge, this work introduces Multiplexed Measurement of Multiple Nuclei (M3N) sequences and proposes MERINA-MP2RAGE, a multi-nuclei sequence, that embeds sodium MRI in a 1H-MP2RAGE acquisition. The developed sequence was implemented on a 7T MRI and tested in phantom and human in vivo experiments. Merging 1H-MP2RAGE and 23Na-MERINA reduces the total scan time by 40% compared to sequential acquisitions while maintaining uncompromised image quality.
|
| Metabolic Processes Studied with Non-Hyperpolarized Other Nuclei (Deuterium)
Henk De Feyter
|
||
0475.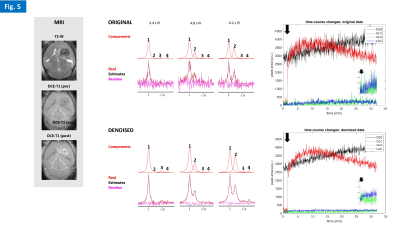 |
Dynamic assessment of early metabolic perturbations in glioma-bearing mice using denoised 2H-MRS
Rui V Simoes1, Javier Istúriz2, Jonas L Olesen3, Sune N Jespersen3, and Noam Shemesh1
1Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 2Neos Biotec, Pamplona, Spain, 3Center of Functionally Integrative Neuroscience (CFIN) and MINDLab, Department of Clinical Medicine, Aarhus University, Aarhus, Denmark
As our understanding of cancer metabolism advances, novel methods are needed to dynamically assess its heterogeneity in preclinical models, with high temporal resolution. Here we show the feasibility of dynamic 2H-MRS in glioma-bearing mice to monitor glucose metabolism with <3s resolution. This was achieved with a new 2H/1H RF coil design for the mouse brain and a novel application of Marchenko–Pastur denoising. We are now adapting this methodology to 2H-MRSI, with high spatial resolution.
|
|
 |
0476.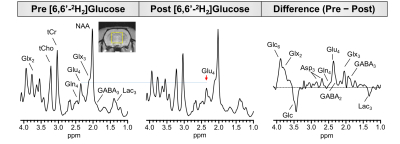 |
Combining 1H MRS with deuterium labeled glucose: A new strategy to assess dynamics of neural metabolism in vivo
Laurie J Rich1, Puneet Bagga1, Gabor Mizsei1, Mitchell D Schnall1, John A Detre2, Mohammad Haris3, and Ravinder Reddy1
1Radiology, University of Pennyslvania, Philadelphia, PA, United States, 2Neurology, University of Pennyslvania, Philadelphia, PA, United States, 3Research Branch, Qatar University, Doha, Qatar
Proton magnetic resonance spectroscopy (1H MRS) is a powerful technique capable of detecting a range of endogenous metabolites, but with currently existing approaches does not enable tracking of metabolic fluxes and pathways. We report a new strategy which utilizes 1H MRS in conjunction with administration of deuterium (2H) labeled glucose to track downstream labeling of neural metabolites. Since 2H is invisible on 1H MRS, replacement of 1H with 2H leads to an overall reduction in 1H MRS signal for the corresponding metabolites. Therefore, this approach makes it possible to monitor neural metabolism using conventional and widely available 1H MRS methodologies.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Video
Watch the Video Back to Top
Back to Top