Digital Poster Session
Spectroscopy:
Spectroscopy
2843 -2857 New Techniques in MRS - MRS Reconstruction & AI
2858 -2873 New Techniques in MRS - MRS Technical 1
2874 -2889 New Techniques in MRS - MRS Technical 2
2890 -2904 New Techniques in MRS - MRS Technical 3
2905 -2920 New Techniques in MRS - MRS Technical 4
2843.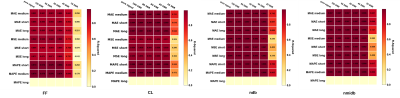 |
Evaluation of Convolutional Neural Network For Magnetic Resonance Spectroscopy of Lipids
Anmol Monga1, Dimitri Martel1, and Gregory Chang1
1Radiology, NYU Langone Health, New York, NY, United States
Recent progress in the convolutional neural network has provided new tools for signal processing and quantification. Magnetic Resonance Spectroscopy is a powerful quantitative tool, which allows notably to access lipid composition. We aim to design and evaluate a neural network for lipid MRS quantification using simulated data.
|
|
2844.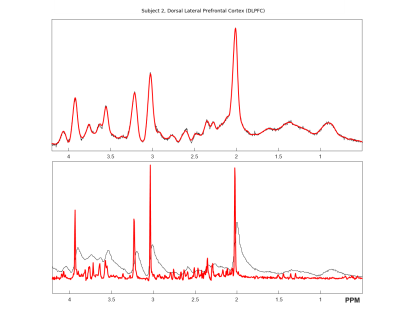 |
Water Scaling Strategy for Metabolites Quantified in MRS by Deep Learning
Yu-Long Huang1, Yi-Ru Lin1, Teng-Yi Huang2, Cheng-Wen Ko3, and Shang-Yueh Tsai4,5
1Department of Electronic and Computer Engineering, National Taiwan University of Science and Technology, Taipei, Taiwan, 2Department of Electrical Engineering, National Taiwan University of Science and Technology, Taipei, Taiwan, 3Department of Computer Science and Engineering, National Sun Yat-Sen University, Kaohsiung, Taiwan, 4Graduate Institute of Applied Physics, National ChengChi University, Taipei, Taiwan, 5Research Center of Mind, Brain and Learning, National ChengChi University, Taipei, Taiwan
Recently, it has been shown that MRS can be analyzed by a convolutional neural network (CNN) with concentrations quantified in a relative way. Here, we propose to scale in vivo MRS data according to water signal in simulated spectra and in vivo data so that CNN spectra can be scaled to institutional units for possible between subject comparison. Our results show that the quantified metabolites are at the same level as those quantified using LCModel with water scaling method but with less repeatability. A further phantom study is necessary to validate the proposed method.
|
|
2845.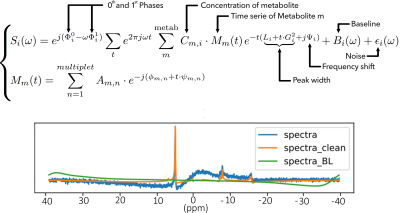 |
31P Magnetic Resonance Spectroscopy analyzed and quantified by Convolutional Neural Network (CNN)
Julien Songeon1, Sebastien Courvoisier1, Antoine Klauser1, Alban Longchamp2, Jean-Marc Corpataux2, Leo Buhler3, and François Lazeyras1
1Department of Radiology and Medical Informatics, University of Geneva, Geneva, Switzerland, 2Department of Vascular Surgery, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland, 3Faculty of Science and Medicine, Section of Medicine, University of Fribourg, Fribourg, Switzerland
Phosphorus magnetic resonance spectroscopy imaging (31P-MRSI) allows the probing of biological compounds that hold fundamental cellular information. High resolution MRSI at 3T suffers from low signal-to-noise ratio (SNR) inherent to the nuclear low sensitivity. This is accentuated in the MRSI in comparison to unlocalized free induction decay (FID) where acquired volumes are smaller and consequently lower the SNR. Our Convolutional Neural Network (CNN) based algorithm perform efficient quantification of metabolite and is compared to last-square fitting algorithm. Our model was trained with a simulated dataset and tested with both simulated spectra and real spectra from 3D 31P-MRSI acquired in kidneys.
|
|
2846.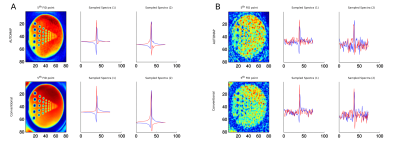 |
MRSI reconstruction pipeline in the age of Deep Learning
Stanislav Motyka1, Lukas Hingerl1, Philipp Moser1, Asan Agibetov2, Georg Dorffner2, and Wolfgang Bogner1
1Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Section for Artificial Intelligence and Decision Support (CeMSIIS), Medical University of Vienna, Vienna, Austria
Whole-brain MRSI measured with a concentric ring trajectories based FID-MRSI sequence generates large amounts of data, which makes post-processing very time-consuming (up to several hours). To speed-up the reconstruction, deep learning approaches could be applied. AUTOMAP provides an attractive solution to reconstruct data directly from non-Cartesian kSpace data. However, it requires single-channel data. Therefore, the coil combination needs to be performed in the kSpace domain. We showed that this strategy is in principle feasible, but requires future work on stability against noise.
|
|
2847.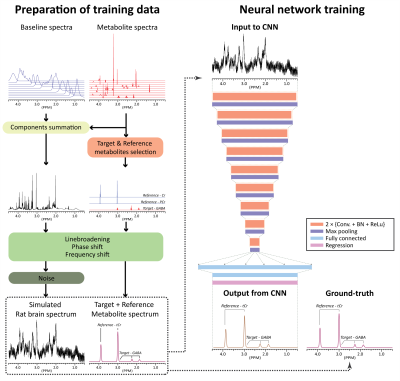 |
Deep learning-based target metabolite-specific signal isolation and big data-driven measurement uncertainty estimation in 1H-MRS of the brain
HyeongHun Lee1 and Hyeonjin Kim1,2
1Department of Biomedical Sciences, Seoul National University, Seoul, Republic of Korea, 2Department of Radiology, Seoul National University Hospital, Seoul, Republic of Korea
We developed convolutional-neural-networks(CNNs) for each individual metabolites capable of spectrally isolating target metabolite signal for quantification on simulated rat brain spectra at 9.4T. Although heuristically and empirically developed, a method of predicting measurement uncertainty is also proposed by exploiting the spectral isolation capability of the CNNs and the availability of big data. The quantitative accuracy of the proposed method was higher than that of the LC model. The measurement uncertainty predicted by the proposed method was highly correlated with the ground-truth error. The proposed method may be used for metabolite quantification with measurement uncertainty estimation in rat brain at 9.4T.
|
|
2848.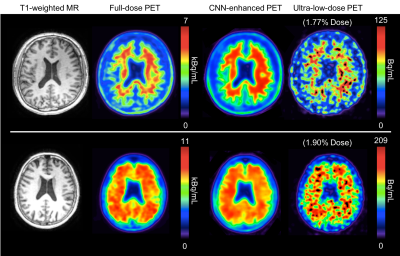 |
MRI-assisted Deep Learning-enhanced Actual Ultra-low-dose Amyloid PET Acquisitions
Kevin T Chen1, Dawn Holley1, Kim Halbert1, Tyler N Toueg1, Athanasia Boumis1, Elizabeth Mormino1, Mehdi Khalighi1, and Greg Zaharchuk1
1Stanford University, Stanford, CA, United States
We are aiming to greatly reduce the radioactive radiotracer dose administered to subjects during PET scanning. In this work we propose to leverage the perfect spatiotemporal correlation of hybrid PET/MRI scanning to synthesize diagnostic PET images from multiple MR images and a noisy PET image reconstructed from acquisitions with actual ultra-low-dose (as low as ~1% of the original) amyloid radiotracer injections, using trained deep neural networks. This technique can potentially increase the utility of hybrid amyloid PET/MR imaging and remove the limiting factors to large-scale clinical longitudinal PET/MRI studies.
|
|
2849.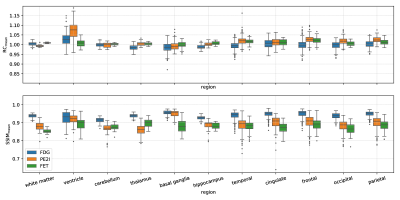 |
Learning MR-guided PET reconstruction in image space using a convolutional neural network - application to multiple PET tracers
Georg Schramm1, David Rigie2, Thomas Vahle3, Ahmadreza Rezaei1, Koen van Laere1, Timothy Shepherd4, Johan Nuyts1, and Fernando Boada2
1Department of Imaging and Pathology, Division of Nuclear Medicine, KU Leuven, Leuven, Belgium, 2Center for Advanced Imaging Innovation and Research (CAI2R) and Bernard and Irene Schwartz Center for Biomedical Imaging, Department of Radiology, New York University School of Medicine, New York City, NY, United States, 3Siemens Healthcare GmbH, Erlangen, Germany, 4Department of Neuroradiology, NYU Langone Health, Department of Radiology, New York University School of Medicine, New York City, NY, United States
Recently, we could show that MR-guided PET reconstruction can be mimicked in image space using a convolutional neural network which facilitates the translation of MR-guided PET reconstructions into clinical routine. In this work, we test the robustness of our CNN against the used input PET tracer. We show that training the CNN with PET images from two different tracers ([18F]FDG and [18F]PE2I), leads to a CNN that also performs very well on a third tracer ([18F]FET) which was not the case when the network was trained on images from one tracer only.
|
|
2850. |
Magnetic Resonance Spectroscopy Denoising with the Automatic Regularization Parameter Estimation in Low-Rank Hankel Matrix Reconstruction
Tianyu Qiu1, Wenjing Liao2, Di Guo3, Zhangren Tu3, Bingwen Hu4, and Xiaobo Qu1
1Department of Electronic Science, Xiamen University, Xiamen, China, 2School of Mathematics, Georgia Institute of Technology, Atlanta, GA, United States, 3School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, 4Department of Physics, East China Normal University, Shanghai, China
In Magnetic Resonance Spectroscopy (MRS), denoising is an important task for MRS due to its poor sensitivity. In this work, noise removal serves as a low rank Hankel matrix reconstruction from noisy free induction decay (FID) signals, since noiseless FID signals are commonly modeled as exponential functions and their Hankel matrices are usually low rank. A faithful denoising mainly depends on the regularization parameter helping to distinguish meaningful signal from noise. We further derived a theoretical bound to automatically set this parameter. Numerical experiments on the realistic MRS data show that noise can be effectively removed with the proposed approach.
|
|
2851.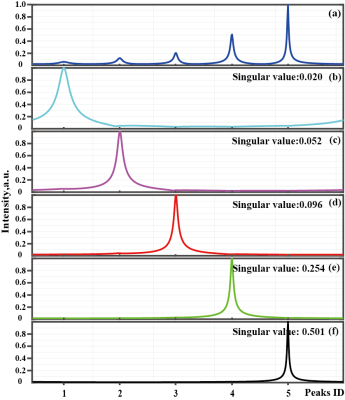 |
A Subspace-based Reliable NMR Spectroscopy Reconstruction
Di Guo1, Zhangren Tu1,2, Tianyu Qiu3, Xiaofeng Du1, Min Xiao2, Vladislav Orekhov4, and Xiaobo Qu3
1School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China, Xiamen, China, 2School of Opto-Electronic and Communication Engineering, Xiamen University of Technology, Xiamen, China, Xiamen, China, 3Department of Electronic Science, Xiamen University, Xiamen, China, Xiamen, China, 4Department of Chemistry and Molecular Biology and Swedish NMR Centre, University of Gothenburg, Gothenburg, Sweden, Gothenburg, Sweden
Accelerating the data acquisition is one of the major developments in modern Nuclear Magnetic Resonance (NMR). Non-Uniform Sampling (NUS) acquires fewer data and reconstructs the spectra with proper signal processing methods1. Here, we introduce an approach to reconstruct faithful spectra from highly accelerated NMR. The FID signal is constrained by the self-learning signal subspace (SLS), in which a true representation of NMR should be in. Results on realistic NMR data demonstrate that the new approach provides much better spectra than the compared state-of-the-art method.
|
|
2852.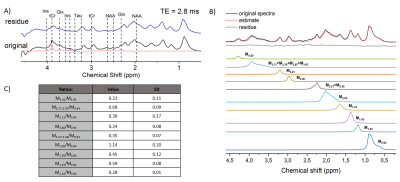 |
Influence of parametrized MM and spline baseline in LCModel using in-vivo rat brain 1H-MR spectra and Monte Carlo simulations at 9.4T
Dunja Simicic1,2, Veronika Rackayova1, Lijing Xin1, Bernard Lanz2, and Cristina Cudalbu1
1CIBM, EPFL, Lausanne, Switzerland, 2LIFMET, EPFL, Lausanne, Switzerland
Reliable detection, post-processing and fitting of MM is crucial for quantifying short-TE 1H-MR brain spectra. In general, an in-vivo acquired full-MM spectrum is included in the basis-set together with a relatively free spline baseline (i.e.LCModel). Availability of high magnetic fields (better resolved in-vivo MM) lead to a need for more sophisticated approaches such as MM parametrization. Furthermore, effect of the stiffness of fitted baseline on the resulting metabolite concentrations gained a lot of interest. We compared parametrized and full-MM basis-sets, with varying DKNTMN to assess the resulting changes in metabolite concentrations (in-vivo rat brain 1H-MRS and Monte-Carlo simulations at 9.4T).
|
|
2853.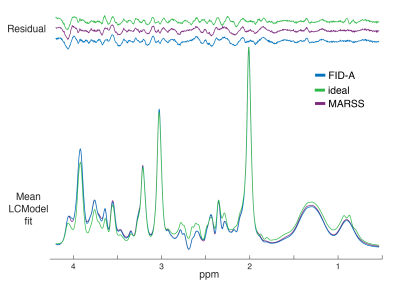 |
Comparison of 3T proton brain spectra quantified with basis sets simulated using experimental and ideal pulses
Michal Povazan1,2, Adam P Berrington3, and Peter B Barker1,2
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University, School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Sir Peter Mansfield Imaging Centre, School of Physics and Astronomy, University of Nottingham, Nottingham, United Kingdom
The most common method for proton MRS quantification is the linear combination of model basis spectra. Typically, these model spectra are simulated using density matrix formalism. Here we aimed to compare the quantification using two basis sets with full localization and real RF pulses and one basis set with non-localized ideal RF pulses. Statistically significant differences were found between metabolite concentrations quantified in LCModel with different basis sets. Whereas two models that used localized real RF pulses were not significantly different for the major metabolites, the ideal basis set differed in concentration for all metabolites except NAA.
|
|
2854.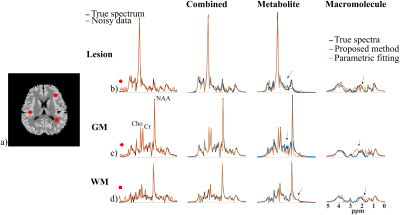 |
Separation of Metabolite and Macromolecule Signals for Short-TE $$$^1$$$H-MRSI Using Learned Nonlinear Models
Yahang Li1,2, Zepeng Wang1,2, Ruo-Jing Ho1,2, and Fan Lam1,2
1Department of Bioengineering, University of Illinois at Urbana-Champaign, URBANA, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, URBANA, IL, United States
We report a novel method to separate metabolite and macromolecule signals from short-TE $$$^1$$$H-MRSI data using learned nonlinear low-dimensional models. A deep-learning-based strategy was developed to learn the nonlinear low-dimensional manifolds where the metabolite and MM signals reside, respectively. A constrained reconstruction formulation is proposed to incorporate the learned model as a prior to reconstruct and separate metabolite and MM signals. The performance of the proposed method was evaluated using both simulation and experimental short TE $$$^1$$$H-MRSI data. Promising results have been obtained, demonstrating the potential of the proposed method in addressing this challenging problem.
|
|
2855.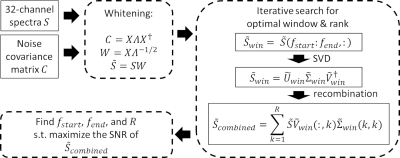 |
Optimized truncation to integrate multi-channel MRS data using rank-R singular value decomposition (OpTIMUS)
Dongsuk Sung1, Benjamin B Risk2, Maame Owusu-Ansah3, Xiaodong Zhong4, Hui Mao3, and Candace Fleischer1,3
1Department of Biomedical Engineering, Georgia Institute of Technology and Emory University, Atlanta, GA, United States, 2Department of Biostatistics and Bioinformatics, Emory University, Atlanta, GA, United States, 3Department of Radiology and Imaging Sciences, Emory University School of Medicine, Atlanta, GA, United States, 4MR R&D Collaborations, Siemens Healthcare, Los Angeles, CA, United States
Multi-channel phased array coils facilitate acquisition of fast, localized, and high signal-to-noise ratio (SNR) magnetic resonance spectroscopy (MRS) data. As individual spectra are acquired from multiple coil channels, it is necessary to combine these data to form a final spectrum. Here, we present an improved approach for combining multi-channel phased array data using spectral windowing followed by a rank-R singular value decomposition (SVD). Our approach, termed ‘OpTIMUS’ was evaluated using SNR and compared to combination methods including whitened SVD (WSVD), S/N2 weighting, and the vendor-supplied reconstruction. OpTIMUS generated the highest SNR across all methods.
|
|
2856.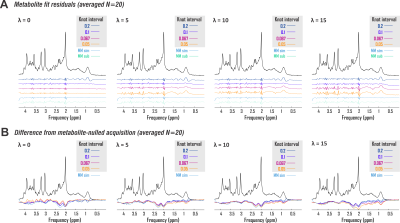 |
Hunting the perfect spline: Baseline handling for accurate macromolecule estimation and metabolite quantification by in vivo 1H MRS
Kelley M. Swanberg1, Karl Landheer1, Martin Gajdosik1, Michael Treacy1, and Christoph Juchem1,2
1Biomedical Engineering, Columbia University School of Engineering and Applied Science, New York, NY, United States, 2Radiology, Columbia University Vagelos College of Physicians and Surgeons, New York, NY, United States
It has been previously established that inappropriately modeled spectral baselines can confound metabolite concentration estimates by in vivo 1H MRS even while contributing to visually reasonable fits. The systematic optimization of spectral quantification pipelines, including baseline modeling, continues nonetheless to be impeded by an under-utilization of validation methods both physically meaningful and involving perfect prior knowledge. Here we address the extant need for evidence-based spectral baseline handling by combining the practical utility of in vivo data with the prior knowledge enabled by simulated standards. We use this paradigm to compare various baseline models for macromolecule characterization and metabolite quantification accuracy.
|
|
2857. |
Comparison between Cartesian and Radial Under-sampling Schemes for Fast MRI using a Deep Cascade of Neural Network
Zhengxin GAO1,2, Zhuoran Jiang2,3, and Zheng Jim Chang2
1Medical Physics Program, Duke Kunshan University, Jiangsu, China, 2Department of Radiation Oncology, Duke Univeristy, Durham, NC, United States, 3Electronic Science and Engineering, Nanjing University, Nanjing, China
Cartesian under-sampling scheme was commonly applied in fast MRI using a deep neural network to simulate the process of fast image acquisition, however, it might not be optimal at a high under-sampling rate. Alternatively, radial under-sampling scheme was used and its efficiency was compared against that of Cartesian under-sampling scheme for T1- or T2-weighted brain, breast, prostate and cervical MRI data at various under-sampling rates. The quantitative evaluation results demonstrated that radial under-sampling scheme could outperformed Cartesian under-sampling scheme on reducing scan time while achieving comparable or better image quality.
|
2858.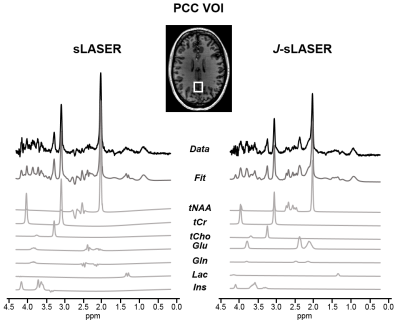 |
J-refocused semi-LASER spectroscopy at 7T: proof-of-concept and application in the human brain at 7T
Chloé Najac1, Vincent Boer2, Hermien E. Kan1, Andrew G. Webb1, and Itamar Ronen1
1C.J. Gorter Center for High Field MRI, Department of Radiology, Leiden University Medical Center, Leiden, Netherlands, 2Danish Research Centre for Magnetic Resonance, Copenhagen University Hospital Hvidovre, Copenhagen, Denmark
Spin-echo sequences suffer from loss of signal due to J-modulation for coupled spins. Previously, it was shown that adding a π/2 pulse between two π pulses in a double spin-echo sequence partially refocuses some of the J-evolution. However, the potential of such sequence at ultrahigh field is limited due to the large chemical shift displacement error. Here, we propose a J-refocused variant of the sLASER sequence (J-sLASER) to improve quantification of J-coupled metabolites at ultrahigh field. Significant improvement in quantitation of J-coupled metabolites is illustrated using simulation, phantom and in vivo measurements in the human brain.
|
|
2859.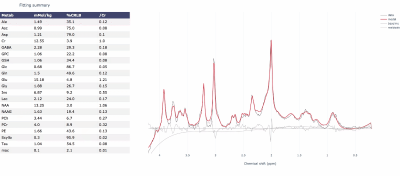 |
FSL-MRS: A New MR Spectroscopy Fitting Tool
William T Clarke1, Jamie Near2, Uzay E Emir3,4, and Saad Jbabdi1
1Wellcome Centre for Integrative Neuroimaging, NDCN, University of Oxford, Oxford, United Kingdom, 2Douglas Mental Health University Institute, and Department of Psychiatry, McGill University, Montreal, QC, Canada, 3School of Health Sciences, Purdue University, West Lafayette, IN, United States, 4Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States
FSL-MRS is a new Python-based MRS fitting tool. The model is based on a linear combination of basis spectra, and fitting is achieved with a sampling approach which gives full posterior distributions of fitted parameters. Results are stored both as text files and an interactive HTML report. On publication the tool will be open source and free as part of the FSL package. The tool will be developed to handle MRSI and model fMRS as time-series. Here we demonstrate an initial implementation in phantom data and three brain regions of 37 subjects and compare against another fitting software.
|
|
2860.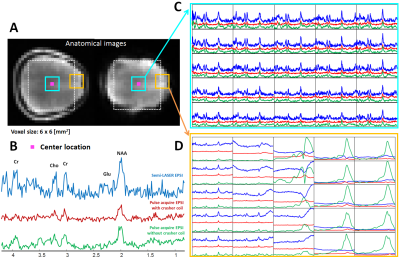 |
Enabling Fast Metabolic Mapping of the Brain using Lipid Crushing and an EPSI Readout at 7T
Kyung Min Nam1, Arjan Hendriks1, Vincent O. Boer2, Dennis D.J. Klomp1, Jannie P. Wijnen1, and Alex Bhogal1
1Imaging & Oncology, UMC Utrecht, Utrecht, Netherlands, 2Danish Research Centre for Magnetic Resonance, Centre for Functional and Diagnostic Imaging and Research, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark
Metabolic mapping at ultra-high field benefits from increased SNR and large chemical shift dispersion, enabling high spatial resolution and more resolved metabolite resonances. By exploiting increased SNR, MRSI acquisitions can be accelerated using echo-planar readout gradients (EPSI) or parallel imaging techniques. However, signal contamination from extra-cranial lipids can degrade the spectral quality and/or introduce fold-over artefacts. Therefore, suppression of extra-cranial lipids during acquisition is imperative for fast spectroscopic imaging. This work aimed to use a “crusher coil” to suppress extracranial lipid signals while acquiring metabolic information using a pulse-acquire acquisition with EPSI readout.
|
|
2861.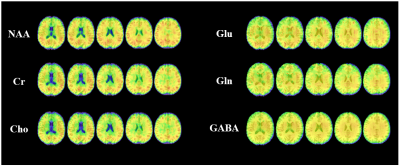 |
Spatiospectral Reconstruction from Hybrid FID/SE J-Resolved MRSI Data with Limited Coverage of (k,t,tJ)-Space
Yibo Zhao1,2, Yudu Li1,2, Jiahui Xiong1,2, Rong Guo1,2, Yao Li3,4, and Zhi-Pei Liang1,2
1Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 4Med-X Research Institute, Shanghai Jiao Tong University, Shanghai, China
J-resolved MRSI using hybrid FID/SE signals has recently been proposed as an effective approach to achieving rapid, high-resolution mapping of brain metabolites and neurotransmitters, an elusive goal of the MRSI community. The new data acquisition scheme poses new problems for data processing, from removal of nuisance signals to reconstruction of spatiospectral distributions. This paper presents an effective method to address these problems, utilizing a union-of-subspaces framework to absorb complementary and prior information. The proposed method has been evaluated using experimental data, producing high-quality spatiospectral distributions of metabolites and neurotransmitters from hybrid FID/SE J-resolved MRSI data with limited coverage of (k,t,tJ)-space.
|
|
2862.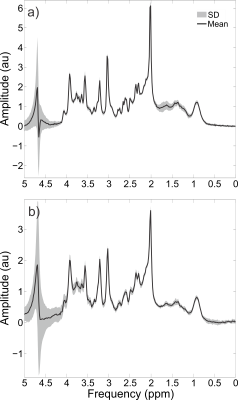 |
Semi-LASER single-voxel spectroscopic sequence with minimal echo time of 20.1 ms: Application in the human brain at 3T
Karl Landheer1, Martin Gajdosik1, and Christoph Juchem1,2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Radiology, Columbia University, New York, NY, United States
The aim of this work was to develop an optimized sLASER sequence which is capable of acquiring artefact-free data with an echo time as short as 20.1 ms on a whole-body clinical 3 T MR system. This was achieved through the use of specialized pulses and optimizing the crusher scheme and phase cycling schemes. High quality spectra were obtained and quantified in 6 healthy volunteers, both in the prefrontal and the occipital cortex.
|
|
2863.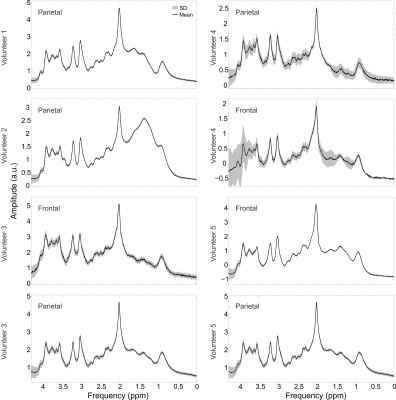 |
UTE-SPECIAL for 3D Localization at an Echo Time of 4 ms on a Clinical 3 Tesla Scanner
Karl Landheer1, Ralph Noeske2, Michael Garwood3, and Christoph Juchem1,4
1Biomedical Engineering, Columbia University, New York, NY, United States, 2GE Healthcare, Berlin, Germany, 3Center for Magnetic Resonance Research and Radiology, University of Minnesota, Twin Cities, MN, United States, 4Radiology, Columbia University, New York, NY, United States
A previously developed magnetic resonance spectroscopy method is improved upon which is now able to obtain spectra with an echo time as short as 4 ms, while recovering the entirety of the available magnetization. This method obtains full 3D spatial localization through a 2D adiabatic inversion pulse which is cycled “on” and “off” every other repetition, in combination with a slice-selective excitation pulse. Both 1D and 2D spectra with an ultrashort echo time of 4 ms are demonstrated at 3T. High quality spectra were obtained for all experiments.
|
|
2864.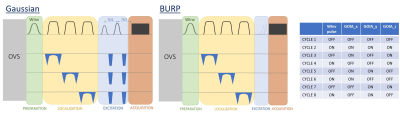 |
A non-water-excitation MRS sequence with zero-echo time to investigate exchangeable moieties in the human brain at 3T
Martyna Dziadosz1, Wolfgang Bogner2, and Roland Kreis1
1Departments of Radiology and Biomedical Research, University of Bern, Bern, Switzerland, 2Department of Biomedical Imaging and Image-guided Therapy, Medical University Vienna, High-field MR Center, Vienna, Austria Most clinical MRS studies concentrate on the upfield part of the spectrum, neglecting the downfield region, since it is mainly composed of labile protons with intensities depending on the water suppression scheme. For reliable and sensitive quantification, a non-water-suppressed or even better a non-water-excitation (NWE) technique is required. We introduce a non-echo technique based on ISIS localization for optimal detection of (moderately) fast exchanging protons at 3T with minimal signal loss due to T2 or exchange and with optimized water magnetization path for longitudinal relaxation enhancement. |
|
2865.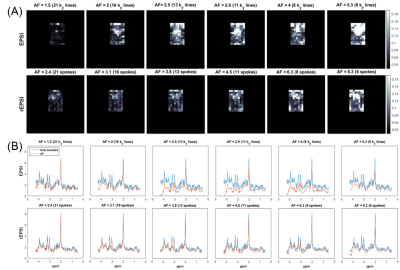 |
Radial Flyback Echo-Planar Spectroscopic Imaging with Golden Angle Sampling and Total Variation-based Compressed Sensing Reconstruction
Andres Saucedo1, Manoj Sarma1, and M. Albert Thomas1
1Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States
In this pilot study, we implement a radial echo-planar spectroscopic imaging (rEPSI) sequence and compare its performance to the Cartesian (EPSI) sequence under multiple rates of retrospective undersampling, for the case of 2D acquisitions in which a single phase-encoded direction is undersampled for EPSI, and a reduced number of radial profiles using golden angle view ordering are reconstructed for rEPSI. Fully sampled data from a healthy volunteer brain and from a brain phantom are acquired. The retrospectively undersampled data is reconstructed using compressed sensing with Total Variation regularization, and the performance of both methods are compared.
|
|
2866.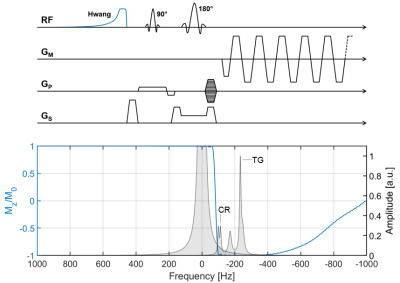 |
Metabolite-cycled echo-planar spectroscopic imaging of the human heart
Sophie M. Peereboom1 and Sebastian Kozerke1
1Institute for Biomedical Engineering, University and ETH Zurich, Zurich, Switzerland
Cardiac triglyceride levels can be assessed using proton MR spectroscopy. While metabolite cycling has already been applied to cardiac proton MRS, the combination of metabolite cycling and spectroscopic imaging for cardiac applications remains to be demonstrated. In this work metabolite-cycled echo-planar spectroscopic imaging is proposed to asses triglyceride-to-water (TG/W) ratios in different regions of the human heart. Results were compared to conventional single-voxel measurements in the interventricular septum. Although SNR is limited for metabolite-cycled EPSI, the method allows for detection of regional TG/W and therefore holds promise to provide insights into regional TG variations.
|
|
2867.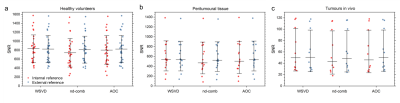 |
Phased-array combination of 2D MRS for lipid composition quantification in patients with breast cancer
Vasiliki Mallikourti1, Sai Man Cheung1, Tanja Gagliardi 2,3, Nicholas Senn1, Yazan Masannat4, Trevor McGordlrick5, Ravi Sharma5, Steven Heys1,4, and Jiabao He1
1Institute of Medical Sciences, School of Medicine, University of Aberdeen, Aberdeen, United Kingdom, 2Department of Clinical Radiology, Aberdeen Royal Infirmary, Aberdeen, United Kingdom, 3Department of Radiology, Royal Marsden Hospital, London, United Kingdom, 4Breast Unit, Aberdeen Royal Infirmary, Aberdeen, United Kingdom, 5Department of Oncology, Aberdeen Royal Infirmary, Aberdeen, United Kingdom
Signal combination algorithms, designed for 1D MRS, were adapted for lipid composition spectra acquired using 2D MRS of double quantum filtered correlation spectroscopy (DQF-COSY). The algorithms of adaptively optimised combination (AOC), noise decorrelated combination (nd-comb), and whitened singular value decomposition (WSVD) were evaluated on lipid composition spectra from healthy volunteers and patients with breast cancer (tumour and peritumoural adipose tissue). WSVD provided maximal SNR uniformly across all spectral peaks. WSVD, through the elimination of additional acquired reference spectrum, shortens acquisition in patients to less than 11 min instead of 17 min while maintaining sensitivity.
|
|
2868.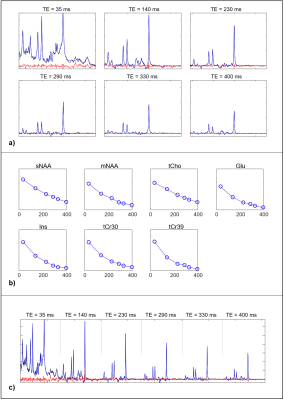 |
Using Simultaneous Multi-echo Fitting to Accelerate Magnetic Resonance Spectroscopic Relaxometry
Patrick J Bolan1, Gregory J Metzger1, Dinesh Deelchand1, and Malgorzata Marjanska1
1Radiology / Center for Magnetic Resonance Research, University of Minnesota-Twin Cities, Minneapolis, MN, United States
Measurements of metabolite T2 relaxation constants can be valuable biomarkers of aging and disease. The conventional method for measuring multiple metabolite T2s is to independently fit spectra from a multi-TE and then separately fit the amplitudes to an exponential decay to estimate T2. In this work we implement a simultaneous fitting approach to fit all of the multi-TE spectra at the same time by incorporating the transverse relaxation in the model. This approach greatly reduces the degrees of freedom and enable T2 estimation in noisy data, which may be used to shorten acquisition times and/or measure smaller regions.
|
|
2869.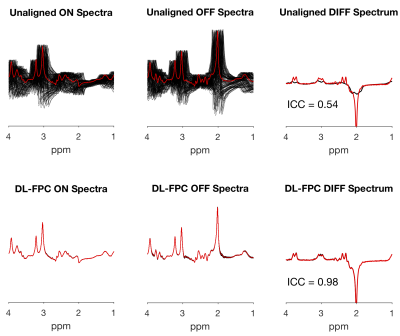 |
Frequency and Phase Correction of J-Difference Edited Spectra using Deep Learning
Sofie Tapper1,2, Mark Mikkelsen1,2, Blake E. Dewey2,3, and Richard A. E. Edden1,2
1Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 3Department of Electrical and Computer Engineering, Johns Hopkins University, Baltimore, MD, United States
Frequency-and-phase correction is an important step in the processing of single-voxel magnetic resonance spectroscopy data, and is required for J-difference editing, which relies on subtraction to reveal a low-SNR signal. We investigated an approach for frequency-and-phase correction using deep learning. Our networks were trained using simulated spectra manipulated with different frequency-and-phase offsets. During validation, the network returned spectra that were corrected to within 1.76 ± 1.19 degrees of phase and 0.09 ± 0.05 Hz of frequency, giving a difference spectrum very similar to the true unmanipulated spectrum. Frequency-and-phase correction is a promising application for deep learning in in vivo MRS.
|
|
2870.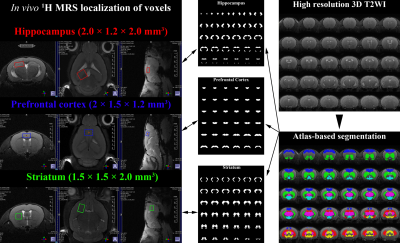 |
Assessment of partial volume effects in MRS voxels applying an Altas-based brain segmentation to high-resolution 3D MRI of rat brain
Chi-Hyeon Yoo1,2, Hyeon-Man Baek2, and Bo-Young Choe1
1The Catholic University of Korea, Seoul, Republic of Korea, 2Gacheon University, Incheon, Republic of Korea
The aim of this study was to apply the atlas-based brain segmentation to high-resolution 3D MRI of the mice brain, to assess a partial volume effect in the voxel. The altas-based segmentation successfully decomposed high-resolution 3D MRI into various anatomical compartments, and by volumetrically analyzing binary masks of the tissue compartment and voxels, a true contribution of the intended tissue compartment in the localized voxel can be assessed. By analyzing the metabolite-specific volume masks, an agreement of the localization between metabolites was evaluated. By evaluating the true contribution of the intended compartment and metabolite-specific agreement, localization reliability can be improved.
|
|
2871.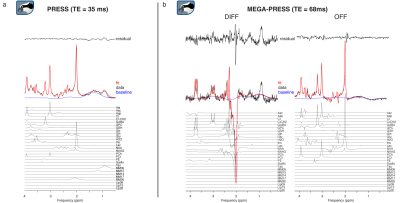 |
Osprey: Open-Source Processing, Reconstruction & Estimation of Magnetic Resonance Spectroscopy Data
Georg Oeltzschner1,2, Helge Jörn Zöllner1,2, and Richard Anthony Edward Edden1,2
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Magnetic resonance spectroscopy (MRS) offers a wide range of methods to study in vivo metabolism. As a result of this diversity, data acquisition and analysis are far from standardized, and researchers frequently develop customized and therefore highly heterogeneous pipelines, often interfacing with commercial or closed-source external fitting software. Here, we present “Osprey”, an all-in-one software package that incorporates all steps of state-of-the-art pre-processing, linear-combination modeling, tissue correction, quantification, and visualization of MRS data from the human brain. Osprey provides a modular, fully open-source framework, allowing the community to rapidly incorporate future methodological developments, accelerate their adaptation, and foster standardization.
|
|
2872.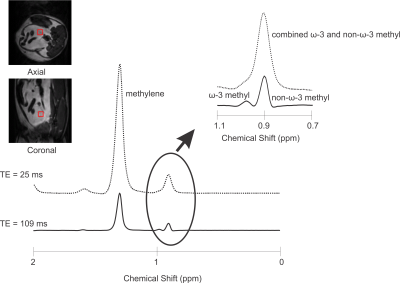 |
Optimized PRESS Echo Time for Quantifying Relative ω-3 Fat Content at 9.4T In Vivo
Clara J. Fallone1, Anthony G. Tessier 1,2, Catherine J. Field3, and Atiyah Yahya1,2
1Department of Oncology, University of Alberta, Edmonton, AB, Canada, 2Department of Medical Physics, Cross Cancer Institute, Edmonton, AB, Canada, 3Department of Agricultural, Food, and Nutritional Science, University of Alberta, Edmonton, AB, Canada
Omega-3 (ω-3) fat content in adipose tissue is relevant to the study of disease. It is challenging to quantify with Magnetic Resonance Spectroscopy (MRS) due to its low concentration in vivo. In addition, its resonance is difficult to resolve from that of the non-ω-3 protons, even at 9.4T. A PRESS (Point RESolved Spectroscopy) echo time of 109ms was determined to yield resolved ω-3 (≈ 0.98ppm) and non-ω-3 (≈ 0.9ppm) methyl peaks at 9.4T. The efficacy of the sequence was verified on oil phantoms and on rats fed a high ω-3 fat diet.
|
|
2873.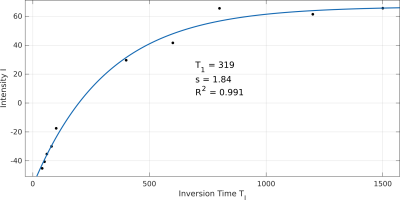 |
In Situ Quantification of the Longitudinal Relaxation Time of Phophatidylcholine in Human Bile at 7T
Lorenz Pfleger1,2, Albrecht Ingo Schmid2,3, Stefan Wampl2,3, Martin Gajdošík1,2,4, Siegfried Trattnig2,5, Michael Krebs1, and Martin Krššák1,2,5
1Division of Endocrinology and Metabolism, Department of Medicine III, Medical University of Vienna, Vienna, Austria, 2High-field MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 3Center for Medical Physics and Biomedical Engineering, Medical University of Vienna, Vienna, Austria, 4Department of Biomedical Engineering, Columbia University, New York, NY, United States, 5Christian Doppler Laboratory for Clinical Molecular MR Imaging, Vienna, Austria
This study focuses on the in vivo determination of the longitudinal relaxation time (T1) of phosphatidylcholine (PtdC) in the gallbladder at 7T and its challenges. In most cases, it was possible to assess T1 with a 1D-ISIS sequence. In cases where the gallbladder is further away from the coil a STEAM sequence, which does not depend on 180° RF pulses, can be used. The T1 values of PtdC ranged between 290 and 480ms.
|
2874. |
Development of absolute quantitation method in 1H MRS at high magnetic field
Hidehiro Watanabe1, Nobuhiro Takaya1, and Fumiyuki Mitsumori1
1Center for Environmental Measurement and Analysis, National Institute for Environmental Studies, Tsukuba, Ibaraki, Japan
The method for absolute quantitation of 1H MRS in a human brain at high B0 field was proposed. A water phantom as a concentration reference and a human brain area measured separately. The ratio of the reception sensitivities between uniform areas in the phantom and in the human brain can be computed from measured B1+s. The ratio between the VOI and the uniform area in the human brain can be computed by our previously reported ratio map method. Then, concentration in the VOI can be calculated. Our method was demonstrated in the phantom experiments.
|
|
2875. |
Development of hydroxyl radical imaging using dynamic nuclear polarization MRI
Shinichi Shoda1, Fuminori Hyodo1, Norikazu Koyasu1, Yoko Tachibana2, Hinako Eto2, and Masayuki Matsuo1
1Gifu University, Gifu, Japan, 2Kyushu University, Fukuoka, Japan
Oxidative stress is implicated in various diseases such as inflammation, neurodegenerative disorders (Alzheimer’s disease, Parkinson’s disease), atherosclerosis, diabetic and cancer. Excess reactive oxygen species (ROS) are produced during altered cellular metabolism in various diseases. Among ROSs, hydroxyl radicals (•OH) is one of the most reactive molecules in biological systems. Therefore, it is considered that monitoring of •OH is could be useful technologies for evaluation of redox mechanism and oxidative diseases. In this study, we developed the hydroxyl radical imaging technique using combining DNP-MRI and DMPO.
|
|
2876.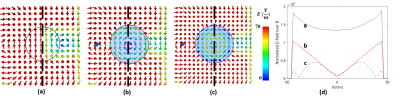 |
Large denoising effect of ultrahigh dielectric constant (uHDC) materials
Navid Pourramzangandji1, Christopher Sica2, Byeong-Yeul Lee3, Hannes Wiesner3, Maryam Sarkarat4, Michael Lanagan4, Xiao-Hong Zhu3, Wei Chen3, and Qing Yang1
1Department of Neurosurgery, PennState University College of Medicine, Hershey, PA, United States, 2Department of Radiology, PennState University College of Medicine, Hershey, PA, United States, 3CMRR, Department of Radiology, University of Minnesota, Minneapolis, MN, United States, 4Department of Engineering Science and Mechanics, Pennsylvania State University, State College, PA, United States
We investigated the effect of ultrahigh dielectric constant (uHDC) materials on reducing the noise and enhancing B1 efficiency at 78-MHz for low-γ nuclear applications (23Na and 13C at 7T or 61-MHz for 17O at 10.5 T). By employing uHDC materials, the conservative E-field can be reduced in the sample, which makes the non-conservative field dominant and increases the transmit efficiency and receive sensitivity. We calculated the normalized noise level and extracted the B1+ efficiency map for baseline (no uHDC) and when uHDC is used, and show experimentally that noise level is reduced considerably in the presence of the UHDC material.
|
|
2877.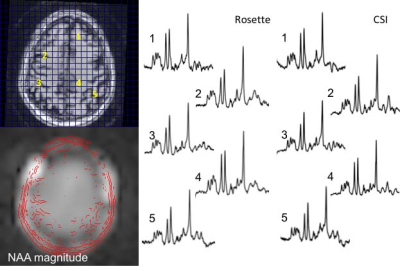 |
Targeted fast spectroscopic imaging at 7T
Jullie W Pan1, Victor Yushmanov1, Chan H Moon1, Frank Lieberman2, and Hoby P Hetherington1
1Radiology, University of Pittsburgh, Pittsburgh, PA, United States, 2Neurology, UPMC, Pittsburgh, PA, United States
The increased SNR at 7T and high efficiency of spatial encoding from fast readout trajectories means that substantially accelerated spectroscopic imaging acquisitions are practical. In this report, we implement targeted multi-slice Hadamard encoded cascaded and simultaneous acquisitions. With the cascaded rosette, minimal CSDE is incurred, generating 4slices in 8min12s. The J-refocused sequence achieves excellent CRLB in <2.3min for a single slice. With a B1 reduction strategy that allows multiple simultaneous Hadamard encoding, the J-refocused sequence is also demonstrated with 4slice acquisitions. As performed in brain tumor patients, the targeted Hadamard rosette is well tolerated and sensitive for detection of pathology.
|
|
2878.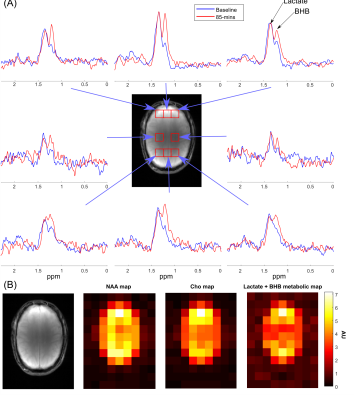 |
Elliptical Localization with Pulsed Second-Order fields (ECLIPSE) for Lactate and BHB Edited Human Brain Proton MRSI
Chathura Kumaragamage1, Henk M De Feyter1, Peter B Brown1, Scott McIntyre1, Terence W Nixon1, and Robin A de Graaf1
1Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States
Βeta-hydroxybutyrate (BHB) and lactate (Lac) are MRS visible metabolites important for brain energy metabolism. In this work we investigate the feasibility to detect elevated levels of BHB with proton MRSI following oral consumption of an energy drink containing 25g of a keto-ester. A gradient-modulated ECLIPSE-OVS method with minimal chemical shift displacement was implemented for lactate and BHB edited MRSI of the human brain. Elevated levels of BHB following oral intake of a keto-ester energy drink were observed in an 8 mL nominal volume MRSI acquired within 9-min at 4 T.
|
|
2879.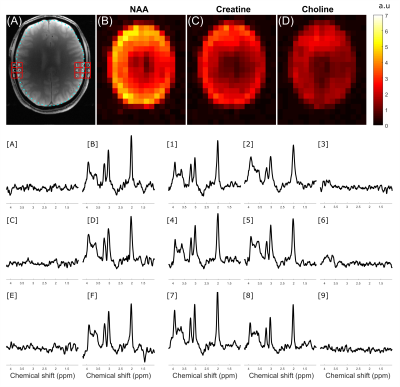 |
ECLIPSE utilizing Gradient Modulated Offset-Independent Adiabaticity (GOIA) Pulses for Human Brain Proton MRSI
Chathura Kumaragamage1, Henk M De Feyter1, Peter B Brown1, Scott McIntyre1, Terence W Nixon1, and Robin A de Graaf1
1Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States
At high magnetic field strengths (≥3T), the utility of MRSI is impeded by challenges such as increased B1 field heterogeneity, increased RF power requirements for reduced chemical shift displacement errors (CSDE), and water/lipid contamination. In this work, the utilization of gradient modulated (GOIA) RF pulses, is investigated, with elliptical localization with pulsed second order fields (ECLIPSE) for MRSI. A 15 kHz GOIA pulse based ECLIPSE double-spin-echo MRSI sequence was developed. In vivo data demonstrate that the GOIA based MRSI method provides full-intensity metabolite spectra in edge of the brain voxels, and undetectable extracranial lipid signals within or outside the brain.
|
|
2880.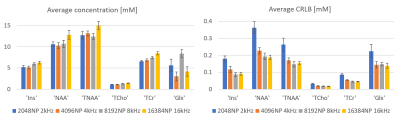 |
The impact of receiver bandwidth and number of points on MRS fitting accuracy
Maria Yanez Lopez1 and Peter J Lally2
1Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Department of Brain Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom
The aim of this work was to investigate the effect of receiver bandwidth and number of points in the fitting of MRS spectra acquired in healthy controls, using a LASER MRS sequence at 3T. Our results highlight the need for harmonisation between different studies in terms of BW/number of points wherever possible, since variations across clinical populations might not be detected due to systematic errors induced from variations in these parameters alone. Optimisation of BW/number of points is also encouraged when setting up a new study.
|
|
2881.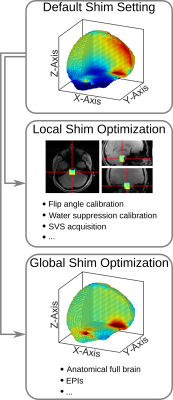 |
Comparison of an advanced, fieldmap-based shimming approach and FASTESTMAP for single-voxel 1H MR spectroscopy at 7T
Ana Gogishvili1,2, Michael Schwerter1,3, Ezequiel Farrher1, Ketevan Kotetishvili2, and N. Jon Shah1,4,5,6
1Institute of Neuroscience and Medicine 4, Foschungszentrum Jülich, Jülich, Germany, 2Engineering Physics Department, Georgian Technical University, Tbilisi, Georgia, 3Faculty of Mathematics, Computer Science and Natural Sciences, RWTH Aachen University, Aachen, Germany, 4Department of Neurology, Faculty of Medicine, RWTH Aachen University, Aachen, Germany, 5JARA - BRAIN - Translational Medicine, RWTH Aachen University, Aachen, Germany, 6Institute of Neuroscience and Medicine 11, Foschungszentrum Jülich, Jülich, Germany
Proton MRS only benefits from ultra-high field strengths if good B0 homogeneity is achieved. For this, projection-based B0 shim techniques, e.g. FASTMAP, are commonly used in single-voxel acquisitions. However, FASTMAP and its variants typically only use up to 2nd order shim coils and only provide shim values for a specific voxel position. In contrast, fieldmap-based approaches for B0 shimming have no shim order restrictions and can be used to calculate multiple shim settings over different ROIs from a single acquisition. Here we compare projection and fieldmap-based shim approaches in brain areas known to be problematic in terms of B0 homogeneity.
|
|
2882.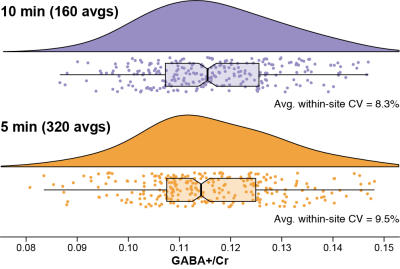 |
Five-minute GABA: Halving the scan time of GABA-edited MRS measurements
Mark Mikkelsen1,2 and Richard A. E. Edden1,2
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Measurement of lower-concentration metabolites at ≤3T using edited MRS requires relatively long scan times (~10 min for a 27-mL voxel). We investigated whether scan times can be halved to ~5 min without a significant increase in group-level variance. GABA-edited MEGA-PRESS data from the Big GABA repository were analyzed by quantifying GABA+/Cr levels for complete datasets (320 averages; ~10 min) versus the first half of each (160 averages; ~5 min). There were no significant differences in group-level variance or GABA+/Cr levels. It appears that for a large, high-SNR voxel, GABA editing scan times of ~5 min are feasible.
|
|
2883.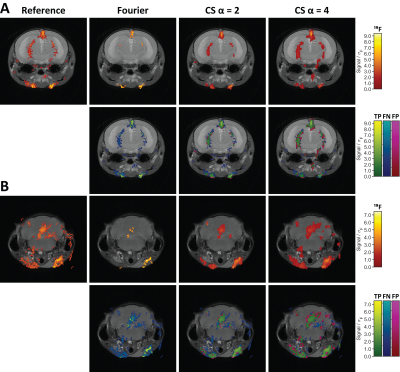 |
Performance of Compressed Sensing for detecting low SNR 19F MRI in the Murine Brain using Prospective Undersampling
Ludger Starke1, Joao dos Santos Periquito1, Christian Prinz1, Andreas Pohlmann1, Thorald Niendorf1,2, and Sonia Waiczies1
1Berlin Ultrahigh Field Facility (B.U.F.F.), Max-Delbrück-Center for Molecular Medicine in the Helmholtz Association, Berlin, Berlin, Germany, 2Charité Campus Buch, Experimental and Clinical Research Center (ECRC), Berlin, Germany
Fluorine-19 (19F) MRI is an established tool for tracking inflammatory cells in vivo due to its excellent detection specificity. Nonetheless, low signal-to-noise ratios remain a major challenge, especially when studying inflammatory cell distribution in the brain. It was shown that compressed sensing (CS) increases 19F MRI sensitivity, yet prospective undersampling using dedicated CS sequences has only been reported in proof of concept experiments. Since false positives were observed in CS reconstructions, this work provides a thorough assessment of detection performance of in vivo CS as a tool for enhancing 19F MRI sensitivity.
|
|
2884.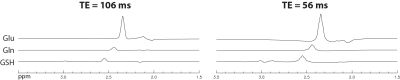 |
Simultaneous Measurement of Glutamate, Glutamine, and Glutathione by Single-Step Spectral Editing
Li An1, Maria Ferraris Araneta1, Milalynn Victorino1, and Jun Shen1
1National Institute of Mental Health, National Institutes of Health, Bethesda, MD, United States
A single-step spectral editing MRS sequence is described for simultaneously measuring glutamate, glutamine, and glutathione at 7 T. Same as a previously described three-step editing technique, the editing pulse induces the collapse of targeted multiplets into high intensity pseudo singlets at a medium echo time. No data subtraction is necessary. In the current example, the editing pulse acts on the H3 protons of glutamate, glutamine, and glutathione near 2.12 ppm to generate sharp and high intensity pseudo singlets of glutamate, glutamine, and glutathione H4 protons. Signal enhancement of the three metabolites is analyzed in detail.
|
|
2885.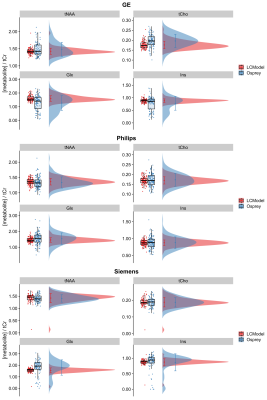 |
Leveraging large publicly available datasets to benchmark new 1H-MRS modeling methods against established algorithms
Helge J. Zöllner1,2, Georg Oeltzschner1,2, Michal Povazan1,2, and Richard A. E. Edden1,2
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Contemporary linear-combination modeling (LCM) methods for MRS data are usually implemented in compiled, ‘black-box’ fashion. The ability to modify underlying algorithms and/or introduce novel quantification approaches may improve the transparency, robustness and accuracy of metabolite estimation, but there is no established framework for rapid prototyping of modeling algorithms and benchmarking their performance. Here, we use a large, publicly available 3T PRESS dataset from multiple sites and vendors to assess the performance of a new open-source LCM algorithm, featured in a new MRS data analysis toolbox (‘Osprey’, github.com/schorschinho/osprey). Quantification of four major metabolites is compared to the widely used LCModel algorithm.
|
|
2886.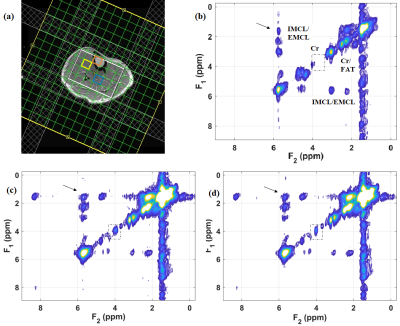 |
Semi LASER Localized Echo Planar Total Correlated Spectroscopic Imaging
Manoj Kumar Sarma1, Andres Saucedo1, Uzay E. Emir2, and M. Albert Thomas1
1Radiological Sciences, David Geffen School of Medicine at UCLA, Los Angeles, CA, United States, 2School of Health Sciences, Purdue University – West Lafayette, West Lafayette, IL, United States
We implemented a semi-LASER localized total correlated spectroscopic imaging (EP-TOCSI) sequence on a 3T MRI scanner. Compared to L-COSY the coherence transfer 900 RF pulse was replaced by a spin-lock module consisting of adiabatic train of RF pulses using MLEV4 phase cycling; also, multi-voxel encoding included EPI read-out combined with phase-encoding. After evaluating the performance of the 4D EP-TOCSI sequence using a corn oil phantom, we investigated calf muscle of 2 healthy and 1 type 2 diabetic subjects. There were relayed cross peaks in addition to COSY cross peaks conventionally recorded by L-COSY or EP-COSI.
|
|
2887.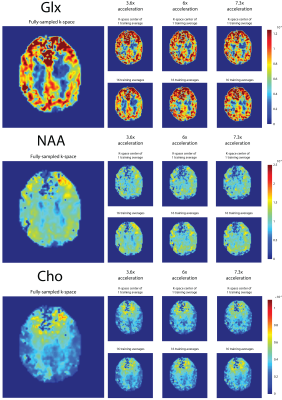 |
Improved MultiNet GRAPPA performance with semi-synthetic calibration data for accelerated 1H FID MRSI at 7T
Kimberly Chan1, Theresia Ziegs2, and Anke Henning1,2
1Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX, United States, 2Max Planck Institute for Biological Cybernetics, Tuebingen, Germany
It has been shown that neural networks combined with variable k-space undersampling (MultiNet GRAPPA) is superior to a conventional GRAPPA reconstruction at 9.4T. Here, the feasibility of performing MultiNet GRAPPA for 1H FID-MRSI at 7T is investigated with and without novel modifications to the original acquisition/reconstruction scheme. In this study, it is shown that MultiNet GRAPPA is shown to be feasible for 1H MRSI acceleration at 7T with a new k-space undersampling scheme for higher signal-to-noise and increased map reliability and use of a novel technique to increase SNR retention using semi-synthetic calibration data without an increase in acquisition time.
|
|
2888.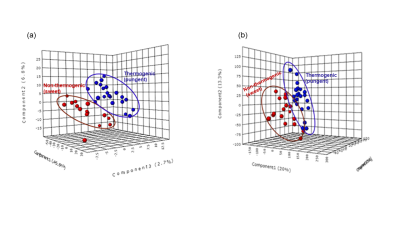 |
Proton NMR metabolomics guided prediction of anti-obesity properties of medicinal plants
Rama Jayasundar1, Aruna Singh1, and Dushyant Kumar1
1Department of NMR, All India Institute of Medical Sciences, New Delhi, India Poster Permission Withheld
Given the alarming increase in obesity and associated NCDs, use of nutritional and medicinal plants in obesity management have come into focus. There is growing interest in pungent thermogenic plants with anti-obesity activity. With this in view, the study has explored the potential of NMR metabolomics to identify plants with anti-obesity activity based on their chemosensory and thermogenic properties, the latter evaluated by Differential Scanning Calorimeter. Multivariate analysis of NMR data discriminated thermogenic (pungent) and non-thermogenic (sweet) plants and also correlated well with the calorimetry analyses. NMR can identify quickly plants with anti-obesity potential using their chemosensory associated thermogenic properties.
|
|
2889.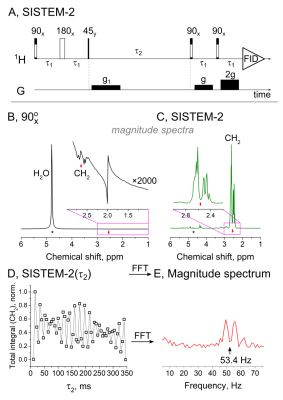 |
Singlet state encoded magnetic resonance (SISTEM) spectroscopy
Andrey Pravdivtsev1, Frank D. Sönnichsen2, and Jan-Bernd Hövener1
1Medical faculty, MOIN CC, Kiel University, Kiel, Germany, 2Otto Diels Institute for Organic Chemistry, Kiel University, Kiel, Germany
MRS provides a non-invasive window into an in vivo biochemistry – basically a virtual biopsy. Here, we present two novel MRS sequences (SISTEM-1 and 2) and method to selectively prepare and detect the signal of endogenous metabolites. The long-lived signal of NAA-CH2 protons was isolated by the strong suppression of H2O and lipid signals using broad-band saturation or double-quantum filter. The frequency of the modulated NAA-CH2 signal was measured and associated with a pH value. Non-invasive measurement of pH value, imaging of drug distribution are only some of the possible applications.
|
2890.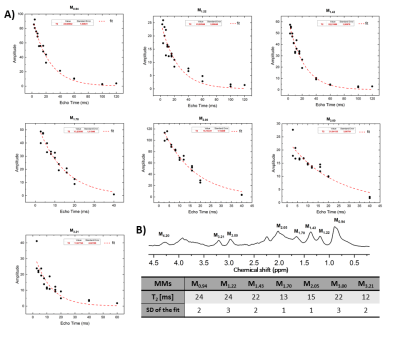 |
T2 relaxation times of seven individual macromolecules in rat brain 1H MR Spectra at 9.4T: single inversion recovery and AMARES post processing
Dunja Simicic1,2, Veronika Rackayova2, and Cristina Cudalbu2
1LIFMET, EPFL, Lausanne, Switzerland, 2CIBM, EPFL, Lausanne, Switzerland
At short echo times 1H-MR spectra contain the contribution of mobile macromolecules (MM), i.e. broader resonances characterized by shorter relaxation times (T1,T2) which underlie the narrower peaks of metabolites. There are very few studies assessing MM T2 relaxation times, with only one study reporting T2-s of individual MM peaks in the full ppm range at 9.4T in the human brain. In this work we present a new approach: single inversion recovery with an optimized inversion time combined with AMARES post-processing. Using this technique we quantified 10-MM components and estimated T2 relaxation times (for 7-MM components) in rat brain at 9.4T.
|
|
2891.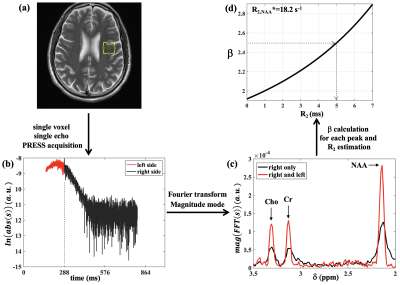 |
Measuring transverse relaxation rates of major brain metabolites from a single spin-echo: Demonstration using single voxel PRESS acquisitions
Reyhaneh Nosrati1,2, Mukund Balasubramanian1,2, and Robert Mulkern1,2
1Radiology, Boston Children's Hospital, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States
Several studies have reported strong correlations between transverse relaxation rate (R2) of the brain metabolites and neurological/psychiatric diseases. Typically, R2 quantification requires several PRESS acquisitions at different echo-times leading to long scan times. Here, we evaluated the feasibility of a novel technique based on a single-echo PRESS acquisition with full echo sampling for R2 estimation of major brain metabolites (NAA, Cr and Cho) and compared those to the values obtained using five PRESS acquisitions. The proposed method allowed for R2 estimation with 87.7±6.8% accuracy compared to the multiple PRESS acquisitions representing a five-fold decrease in acquisition time in this case.
|
|
2892.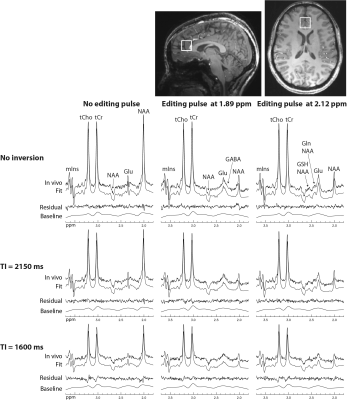 |
Determination of Brain Metabolite T1 Without Interference from Macromolecule Signals
Li An1, Maria Ferraris Araneta1, Milalynn Victorino1, and Jun Shen1
1National Institute of Mental Health, National Institutes of Health, Bethesda, MD, United States
Optimal detection of many metabolites requires a specific TE, at which significant macromolecule signals are often present. The presence of macromolecule signals complicates the measurement of metabolite T1 because different spectral models may be necessary for macromolecules at different inversion recovery stage, therefore, potentially introducing additional errors. To minimize interference from macromolecules, we chose inversion times in such a way that the metabolite signals were changed by as much as 60% while the macromolecule signals were essentially unchanged. This avoided the T1 relaxation effect of macromolecules, and thus simplified data acquisition and post-processing.
|
|
2893.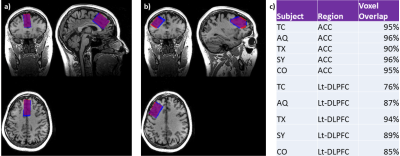 |
Test-Retest Reproducibility of in vivo Cortical GABA and Glx Measurements with MEGA-PRESS: Comparing 32-Channel and 8-Channel Head Coils
Peter Truong1, Napapon Sailasuta1,2, and Sofia Chavez1,2
1Research Imaging Centre - MRI, Centre for Addiction and Mental Health, Toronto, ON, Canada, 2Psychiatry, University of Toronto, Toronto, ON, Canada
Higher-numbered phased-array coils have been associated with significant increases in SNR. For this gain, one would expect more accurate and precise MRS measurements for a regular PRESS sequence processed with a fully automated program such as LCModel. However, the effects of increased SNR on a J-edited technique such as MEGA-PRESS for GABA measurements are not as straight-forward. In this study, we wish to compare the reproducibility of GABA+ and Glx measurements from an 8-channel and a 32-channel head coil in two cortical regions of interest: Anterior Cingulate Cortex (ACC) and Left-Dorsolateral Prefrontal Cortex (Lt-DLPFC).
|
|
2894.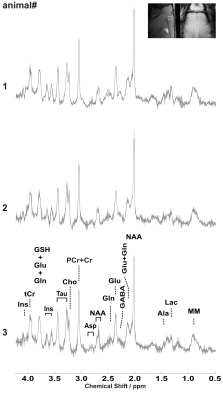 |
Reproducibility of neurochemicals in sub-microlitre MRS
Alireza Abaei1, Dinesh K Deelchand2, Francesco Roselli3, and Volker Rasche1
1Core Facility Small Animal Imaging, Ulm University, Medical Center, ulm, Germany, 2Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States, 3German Center for Neurodegenerative Diseases (DZNE), ulm, Germany
Using sub-microlitre voxel size in magnetic resonance spectroscopy (MRS) is advantageous to investigate specific small brain region without any partial volume effect. The aim of this study was to determine the intra-individual coefficient-of-variation of neurochemical concentrations when using sub-microlitre MRS voxel measured during 3 consecutive days. This study was performed in adult mice at 11.7T. The intra-individual CV for the major and high concentration brain metabolites was ≤ 9% for MRS data collected in pre-clinically feasible scan times. This in turn shows that sub-microlitre can be used in pre-clinical application to detect subtle changes in neurochemical profile in diseased brain.
|
|
2895.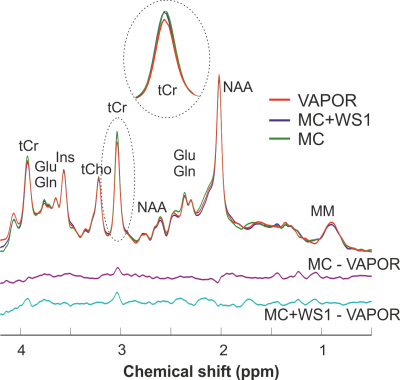 |
Effect of water suppression and metabolite cycling on quantification of 1H MRS spectra in the human brain at 3 Tesla
Dinesh K Deelchand1 and Pierre-Gilles Henry1
1Center for Magnetic Resonance Research, Radiology, University of Minnesota, Minneapolis, MN, United States
This study investigates the effect of water suppression and metabolite cycling on metabolite quantification and macromolecules in 1H MRS. Single-voxel semi-LASER spectra and metabolite-nulled spectra (macromolecules) were acquired in the posterior cingulate cortex in the human brain at 3T using three different schemes: VAPOR, metabolite-cycling (MC), and metabolite-cycling combined with a single water suppression pulse. Results show that macromolecule signal and several metabolites (ascorbate, total creatine, glutamate, and glutathione) have higher in concentration when using MC compared to VAPOR. Combining MC with a single water suppression pulse results in concentrations in between those obtained with VAPOR and MC.
|
|
2896.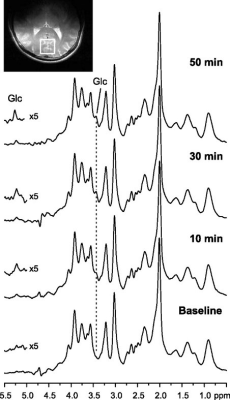 |
An Untargeted Metabolomics Approach to MRS in the Human Brain: A Comparison between LCModel and MRS-based Classifier-Development System
Srijyotsna Volety1, Elizabeth Seaquist2, Gulin Oz3, and Uzay Emir1,4
1Health Sciences, Purdue University, West Lafayette, IN, United States, 2Department of Medicine, Medical School, University of Minnesota, Minneapolis, MN, United States, 3Center for Magnetic Resonance Research, Department of Radiology, University of Minnesota, Minneapolis, MN, United States, 4Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States
MRS quantification tools such as LCModel require the use of prior knowledge and can be time-consuming. Therefore, we propose an untargeted metabolomics approach to in vivo 1H-MRS using pattern recognition and machine learning to analyze spectral features. The ability of our approach to measure changes in brain glucose while blood glucose levels were increased was studied using high quality spectra and reliable data acquisition methods. Results showed similar time-course glucose signals and sensitivity to changes in glucose concentrations for both LCModel and our pattern recognition analysis. Thus, demonstrating that untargeted metabolomics techniques can be used for in vivo MRS quantification.
|
|
2897.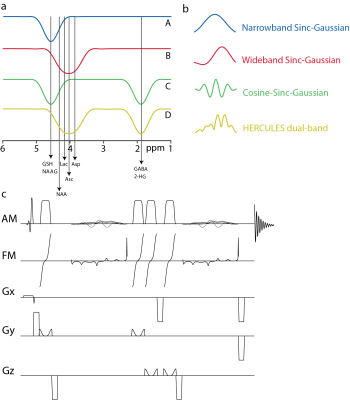 |
MODIFIED HERCULES MULTI-METABOLITE EDITING with SEMI-LASER LOCALIZATION at 3T
Steve CN Hui1,2, Muhammad G Saleh1,2, Georg Oeltzschner1,2, Mark Mikkelsen1,2, Sofie Tapper1,2, and Richard AE Edden1,2
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
An improved editing scheme was proposed for the Hadamard Editing Resolves Chemicals Using Linear-combination Estimation of Spectra (HERCULES). The original editing pulse at 4.18 ppm was replaced by a new wider-band pulse at 4.04 ppm to facilitate signal acquisition for aspartate, ascorbate and lactate. Furthermore, the new scheme (HERCULES-2) was implemented within the semi-LASER and PRESS sequences. HERCULES-2 with semi-LASER yielded increases in overall signal intensities in most targeted metabolites, robust to B1 inhomogeneity, facilitating the precise quantification of edited spectra.
|
|
2898.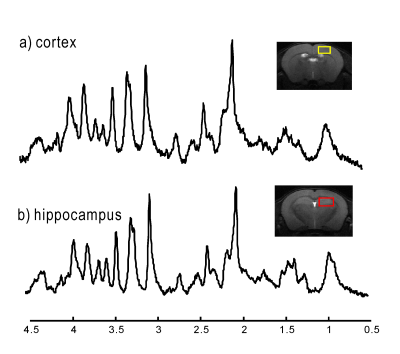 |
Dual-voxel 1H MR spectroscopy in mouse brain at 14.1T with dynamic shim update (DSU) acquisition scheme
Hikari Yoshihara1, Nicolas Kunz2, Jean-Claude Martinou3, and Hongxia Lei2
1Laboratory for Metabolic Imaging, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 2Animal Imaging and Technology Core (AIT), Center for Biomedical Imaging (CIBM), Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 3Faculty of Science, University of Geneva, Geneva, Switzerland Poster Permission Withheld
Dynamic shimming updates with first- and second-order shims are essential for dual-voxel spectroscopy of mouse cortex and hippocampus at high magnetic field. We show that DSU is feasible for studying pharmacological effects in transgenic mice at 14.1 T.
|
|
2899.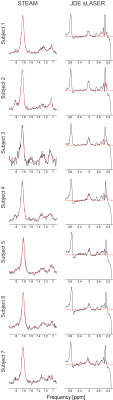 |
A method to estimate GABA free from homocarnosine and macromolecular overlap through a combination of STEAM and J-difference editing sLASER at 7T
Karl Landheer1, Hetty Prinsen2, Ognen A Petroff3, Douglas L Rothman2, and Christoph Juchem1,4
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States, 3Neurology, Yale University, New Haven, CT, United States, 4Radiology, Columbia University, New York, NY, United States
The aim of this work was to develop a novel method to assess accurately both the relative concentrations of homocarnosine as well as GABA free from overlapping creatine, homocarnosine and macromolecule signal. This was achieved via the combination of short echo time STEAM and MEGA-sLASER experiments at 7T. The metabolites GABA and homocarnosine were measured in 6 healthy control subjects, and in a single subject medicated with isoniazid. It was found that (16.6 ± 10.2)% of the supposed GABA signal originated from homocarnosine, and that isoniazid caused a significantly elevated concentration of GABA and homocarnosine in a single subject.
|
|
2900.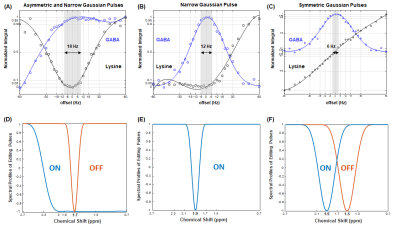 |
Improved GABA editing and macromolecule suppression with adiabatic and highly selective Gaussian pulses at 7T
Song-I Lim1 and Lijing Xin1
1Centre d'Imagerie BioMédicale (CIBM), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
The aim of the study is to improve the efficiency of GABA editing and macromolecule suppression at 7T. An asymmetric adiabatic pulse with broad inversion bandwidth was applied at 1.9 ppm to invert 1.5 – 1.9 ppm range and a narrow Gaussian pulse with 95 Hz of bandwidth (0.95 < Mz) was applied at 1.7 ppm to suppress macromolecule. In the GABA and lysine phantom tests, this scheme shows ± 9 Hz of signal loss threshold in comparison with highly selective inversion pulse without macromolecule suppression (± 6 Hz of threshold) and symmetric pulses (± 3 Hz of threshold).
|
|
2901.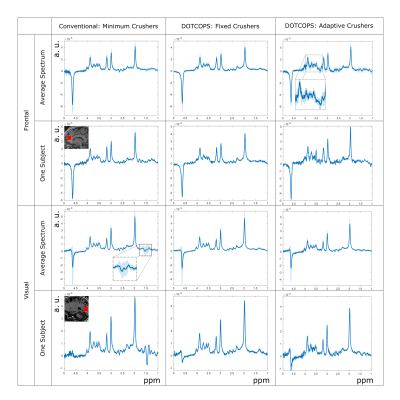 |
Optimal and efficient crushing for semi-LASER at 7T – a comparison
Zahra Shams1, Vincent Oltman Boer2, Dennis W.J. Klomp1, Jannie P. Wijnen1, and Evita Wiegers1
1Radiology, University Medical Center Utrecht, Utrecht, Netherlands, 2Danish Research Centre for Magnetic Resonance, Centre for Functional and Diagnostic Imaging and Research, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark
This study compares different crushing schemes for short-TE semi-LASER at 7T; minimal crushers, DOTCOPS and newly designed Adaptive crusher scheme. Maintaining a short echo time is traded for optimal spoiling of unwanted coherences.
|
|
2902.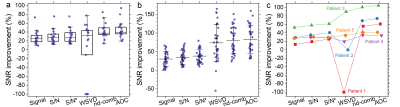 |
Optimal phased-array signal combination for polyunsaturated fatty acids measurement in breast cancer using MQC MR Spectroscopy at 3T
Vasiliki Mallikourti1, Sai Man Cheung1, Tanja Gagliardi 2,3, Yazan Masannat4, Steven Heys1,4, and Jiabao He1
1Institute of Medical Sciences, School of Medicine, University of Aberdeen, Aberdeen, United Kingdom, 2Department of Clinical Radiology, Aberdeen Royal Infirmary, Aberdeen, United Kingdom, 3Department of Radiology, Royal Marsden Hospital, London, United Kingdom, 4Breast Unit, Aberdeen Royal Infirmary, Aberdeen, United Kingdom
Phased-array signal combination algorithms of adaptively optimised combination (AOC), noise decorrelated combination (nd-comb), whitened singular value decomposition (WSVD), S/N2, S/N, and Signal Weighting, were evaluated for enhancing the SNR of polyunsaturated fatty acids spectra acquired using MQC MRS from breast cancer. Data were acquired from seventeen breast tumours excised from patients, fifteen healthy volunteers and five patients with breast cancer. AOC yielded the maximum SNR improvement and was independent from voxel volume, PUFA content, or water/fat ratio. AOC improved SNR by 24% (relative to non-noise decorrelated algorithms) and shortened acquisition time in patients by 1.5 times while maintaining SNR.
|
|
2903.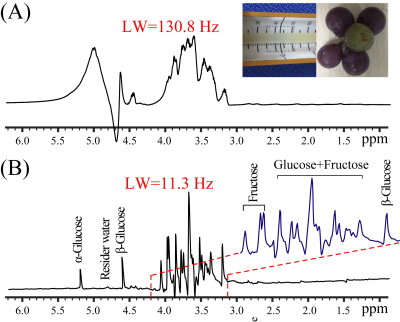 |
High-resolution measurements on heterogeneous samples by spatially-selective pure shift NMR spectroscopy
Yuqing Huang1, Haolin Zhan1, and Zhong Chen1
1Department of Electronic Science, Xiamen University, Xiamen, China
Measurements on heterogeneous samples, such as semisolid food and biological tissues, constitute a significant research topic in various fields. However, measurements on heterogeneous samples by conventional 1D 1H NMR generally suffer from two challenges, namely magnetic field inhomogeneity caused by anisotropic interactions and spectral congestion induced by crowded NMR resonances. In this report, we introduce a spatially-selective pure shift NMR approach for high-resolution measurements on heterogeneous samples based on the suppression on effects of magnetic field inhomogeneity and J couplings, thus useful for investigating heterogeneous sample systems with extensive chemical and biological applications.
|
|
2904.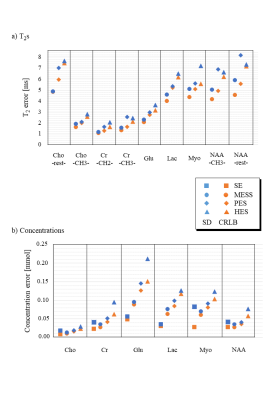 |
Potential benefits from Multi-Echo Single-Shot Spectroscopy with a combined fitting process
Rudy Rizzo1 and Roland Kreis1
1Department of Radiology and Biomedical Research, University of Bern, Bern, Switzerland
The benefits of multi-echo single-shot (MESS) spectroscopy are explored aiming at simultaneous determination of metabolite content and T2 times through simultaneous linear-combination model fitting of partially sampled echoes. Cramer-Rao lower bounds (CRLB) and Monte-Carlo simulations are used to judge this benefit. The novel scheme was compared with traditional multi-echo multi-shot and single-echo methods, exploring different TE settings for spectra of the major brain metabolites. Results indicate that MESS outperforms older methods for simultaneous determinations of T2s and concentrations, with improvements ranging at 20-30% for T2s and 30-50% for areas. However, for concentrations alone traditional single-echo sequences are more sensitive.
|
2905.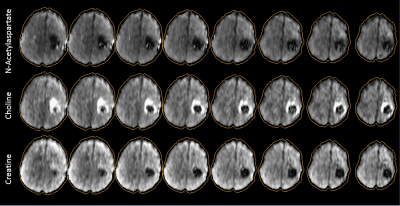 |
High-Resolution Whole-Brain 3D Magnetic Resonance Spectroscopic Imaging
Mohammed Goryawala1, Sulaiman Sheriff1, Ronald Ouwerkerk2, Hari Hariharan3, Peter Barker4, Hyunsuk Shim5, and Andrew Maudsley1
1Radiology, University of Miami, Miami, FL, United States, 2Biomedical and Metabolic Imaging Branch, The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), Bethesda, MD, United States, 3Center for Magnetic Resonance & Optical Imaging, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 4Department of Radiology and Oncology, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 5Department of Radiation Oncology, Emory University School of Medicine, Atlanta, GA, United States
Whole-brain Magnetic Resonance Spectroscopic Imaging (MRSI) is an effective technique for non-invasive quantification of brain metabolite levels 1-3 that can be used to create maps for the study of both regional or diffuse metabolic alterations in various pathologies. Improved spatial resolution can result in significantly better mapping but is often limited by the signal-to-noise ratio (SNR). This abstract presents a whole-brain 3D MRSI acquisition scheme that uses hypergeometric dual-band (HGDB) pulses for lipid suppression over the brain volume, real-time frequency drift correction, and novel post-processing methods to generate whole-brain metabolite maps in humans at 3T.
|
|
2906.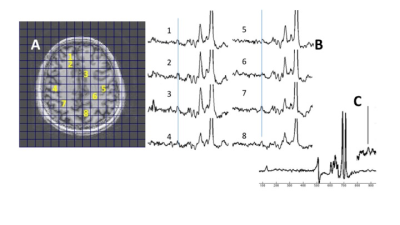 |
Fast spectroscopic imaging of GABA at 7T
Jullie W Pan1, Victor Yushmanov1, Chan H Moon1, and Hoby P Hetherington1
1Radiology, University of Pittsburgh, Pittsburgh, PA, United States
We have developed and implemented a fast spectroscopic imaging acquisition for selective homonuclear polarization transfer sequence for the detection of GABA at 7T. With the high SNR available at 7T, whole slice studies of 1.65cc nominal resolution are achievable with acquisition durations (5 to 10min) conventionally used in single voxel studies. As this editing is performed with longitudinal magnetization, this method is single shot and is flexible for evaluation of multiple coupled spins.
|
|
2907.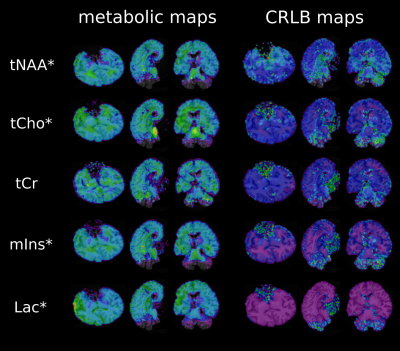 |
Ex vivo whole brain metabolic fingerprinting at 7T - protocol optimization and preliminary results
Alexandra Lipka1,2, Lukas Hingerl1, Gilbert Hangel1, Eva Heckova1, Stanislav Motyka1, Verena Endmayr3, Romana Höftberger3, Simon Hametner4, Siegfried Trattnig1,2, and Wolfgang Bogner1,2
1Highfield MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Christian Doppler Laboratory for Clinical Molecular MR Imaging, Vienna, Austria, 3Institute for Neurology, Medical University of Vienna, Vienna, Austria, 4Center for Brain Research, Institute of Neuroimmunology, Medical University of Vienna, Vienna, Austria MRS has rarely been applied for postmortem measurements except for single-voxel studies, which can be explained by the fact that rapid degradation of metabolite levels makes scanning of fresh specimen highly critical.SVS contrary to MRSI cannot cover whole affected regions, while pathological changes may not be visible on anatomical images, yet. To investigate changes ex-vivo, a true whole-brain MRSI approach is critical. Our study shows the potential of ultra-highfield whole-brain FID-MRSI for metabolic fingerprinting of fresh postmortem brains for correlation with histology, as the link between neurochemical changes and underlying cell-types in many demyelinating/neurodegenerative disorders is still not fully understood. |
|
2908.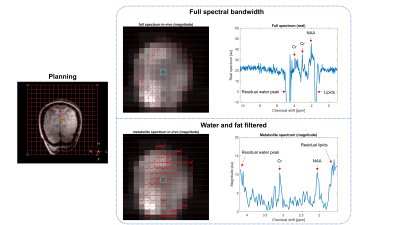 |
Silent EPSI using a gradient axis driven at 20 kHz
Edwin Versteeg1, Kyungmin Nam1, Dennis Klomp1, Jeroen Hendrikse1, Alex Bhogal1, Jeroen Siero1,2, and Jannie Wijnen1
1Radiology, University Medical Center Utrecht, Utrecht, Netherlands, 2Spinoza Centre for Neuroimaging Amsterdam, Amsterdam, Netherlands
Echo planar spectroscopic imaging (EPSI) can be used for fast spectroscopic imaging, however, the gradient hardware greatly limits the spectral bandwidth. Moreover, the associated fast switching gradient produces uncomfortable levels of acoustic noise. In this work, we show the feasibility of spectroscopic imaging at high spectral bandwidth using a silent gradient axis driven at 20 kHz and a segmented EPSI sequence. We report results on the silent segmented EPSI compared to the conventional EPSI approach on a phantom containing NAA, creatine and glutamate and show the first silent and high bandwidth in-vivo measurements.
|
|
2909.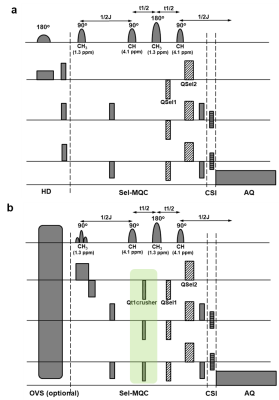 |
Improvement of the selective multiple quantum coherence chemical shift imaging sequence for lactate imaging of lymphoma patients at 3T
Seung-Cheol Lee1, Hari Hariharan1, Fernando Arias-Mendoza1, Gabor Mizsei1, Kavindra Nath1, Sanjeev Chawla1, Stephen J. Schuster1, Ravinder Reddy1, and Jerry Glickson1
1University of Pennsylvania, Philadelphia, PA, United States
Lactate is an important marker of tumor metabolism. We have previously reported the Hadamard slice selected, selective multiple quantum coherence chemical shift imaging sequence for a 3T clinical scanner to image lactate in the human body. Here we report modification and optimization of the sequence for reducing motion effect and increasing lipid suppression while retaining lactate. A coherence selection gradient ratio of a:-1:2 with nonzero a provided improved suppression of lipid resonances at 1.3 ppm and 0.9 ppm compared to the previous 0:-1:2 ratio. Optimization of RF pulses blocked occurrence of MQC between lipid 2.0 ppm and 5.3 ppm resonances.
|
|
2910.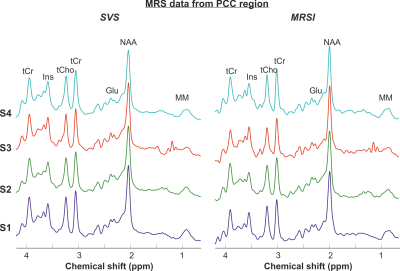 |
Comparison of short echo-time spiral MRSI and single-voxel spectroscopy in the human brain at 3T
Dinesh K Deelchand1 and Pierre-Gilles Henry1
1Center for Magnetic Resonance Research, Radiology, University of Minnesota, Minneapolis, MN, United States
The goal of this study was to compare the spectral quality and metabolite concentrations obtained with short echo-time 2D spiral MRSI and single-voxel spectroscopy (SVS) in the human brain at 3T. Semi-LASER was used for localization (SVS) or pre-localization (MRSI). MRSI was acquired in a transverse slice through the posterior cingulate cortex (PCC), and SVS was from PCC. Results show comparable spectral quality between acquisitions. Differences in concentrations between the two techniques is likely due to the MRSI point-spread function. This study shows feasibility of acquiring high-quality spiral MRSI data similar in spectral quality to SVS within a few minutes.
|
|
2911.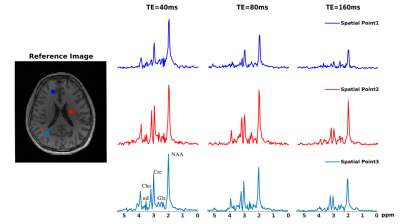 |
B0 Inhomogeneity Corrected Reconstruction for Low-Resolution J-resolved MRSI Using Low-Rank and Spatial Constraints
Zepeng Wang1,2 and Fan Lam1,2
1Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States
This work presents a new method for B0 inhomogeneitycorrected reconstruction of J-resolved (multi-TE) MRSI datawith limited k-space coverage. A constrained reconstruction formulation was proposed to simultaneously reconstruct the data at all TEs, using a joint low-rank constraint that exploits the spatial-spectral-TE correlation in the data and a spatial constraint that exploits prior edge information from high-resolution anatomical images. Simulation and in vivo experiments using a customized 3D J-resolved EPSI acquisition have been performed to evaluate the proposed method. Experimental results demonstrated improved lineshape and SNR achieved by the proposed method over existing B0 correction methods.
|
|
2912.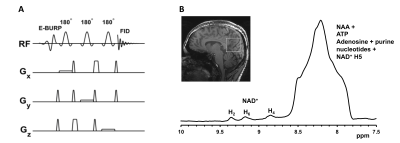 |
3D CSI downfield 1H MR spectroscopy using EBURP1 spectrally selective excitation pulse
Puneet Bagga1, Hari Hariharan1, Walter R Witschey1, and Ravinder Reddy1
1Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States
Downfield MRS provides a great tool to detect and quantify metabolites downfield to water including NAD and ATP. Acquisition of downfield MR spectrum in vivo without water suppression leads to the possibility of detection of many cross-relaxing/exchanging resonances. The T2 of many species occurring downfield of water are relatively short which require short echo time MRS methods. EBURP pulses have been shown to reliably excite narrow spectral region with minimum phase response. In this study, we generated new pulse sequences for single voxel and 3D spectral selective EBURP excitation to detect cerebral NAD from human brain at 7T.
|
|
2913.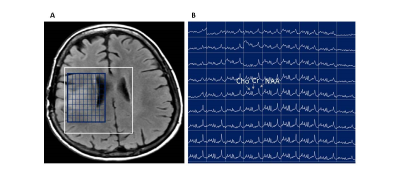 |
Rapid acquisition of semi-LASER MR spectroscopic imaging using symmetric EPSI encoding: A preliminary study in phantom and human brain
Sunitha Thakur1, Olivia Sutton1, Eduardo Coello2, Alexander Lin2, Ralph Noeske3, Ricardo Otazo1, and Robert Young1
1Memorial Sloan Kettering Cancer Center, New York, NY, United States, 2Brigham and Women’s Hospital, Boston, MA, United States, 3MR Applied Science Lab, GE Healthcare, Berlin, Germany
The aim of this study was to develop and test rapid MR spectroscopic imaging (MRSI) in patients with brain tumors. We implemented and optimized rapid MRSI at 3T using semi-localization by adiabatic selective refocusing pulses (semi-LASER), fast spatial encoding using symmetric echo planar spectroscopic imaging (EPSI), and robust water suppression with variable power and optimized relaxation delays. This rapid imaging approach was tested in a phantom and subsequently in a volunteer and a patient. In vivo studies demonstrated that high quality MRSI data with voxel volumes less than 0.5 mL can be collected within a clinically feasible time (2 minutes).
|
|
2914.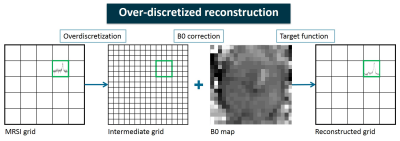 |
Over-discretized reconstruction to correct for B0 inhomogeneity and improve localization in 1H-MRSI of the prostate
Carlijn Tenbergen1, Loreen Ruhm2, Arend Heerschap1, Anke Henning2,3, and Tom Scheenen1
1Radiology and Nuclear Medicine, Radboud University Medical Center, Nijmegen, Netherlands, 2High-Field MR Center, Max Planck Institute for Biological Cybernetics, Tübingen, Germany, 3Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX, United States
In in vivo MRSI spatial inhomogeneities of the main magnetic field cause spectral line broadening and location dependent frequency shifts in the spectra. An over-discretized reconstruction method is applied to existing prostate 1H-MRSI data, consisting of spatial over-discretization of the MRSI grid, frequency shift correction according to a high resolution B0map and weighted spatial averaging to the target resolution. Intravoxel B0 variations are corrected for and resampling to a new target resolution results in improved localization of signals. This could lead to detection of signals with lower spectral intensity and to improved visualization of smaller cancer lesions in the prostate.
|
|
2915.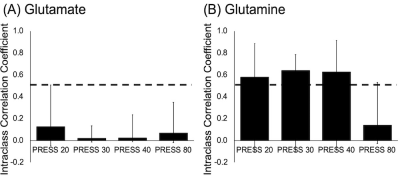 |
Examination of methods to optimize Glutamate-Glutamine separation at 3T
Tiffany Bell1,2,3, Dana Goerzen4, Jamie Near4, and Ashley D Harris1,2,3
1Department of Radiology, University of Calgary, Calgary, AB, Canada, 2Hotchkiss Brain Institute, Calgary, AB, Canada, 3Alberta Children's Hospital Research Institute, Calgary, AB, Canada, 4Douglas Mental Health Institute, Centre d’Imagerie Cerebrale, Montreal, QC, Canada
Separating glutamate (Glu) from its precursor, glutamine (Gln) at 3T is challenging due to signal overlap. Multiple sequence optimisations have been suggested, with simulated data used to assess accuracy. Here, using 7T data as a reference, we investigate the ability of four PRESS sequences with different echo times of separating Glu and Gln. We found that Glu measured at 3T using TE=20ms shows the highest agreement with 7T data. Gln measured using TE=20ms, 30ms and 40ms all agree with Gln measured at 7T. Therefore, we conclude PRESS with TE=20ms is the most effective at separating and quantifying Glu and Gln.
|
|
2916.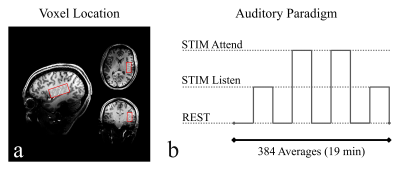 |
Reproducibility of Auditory Functional Magnetic Resonance Spectroscopy at 7T
Eduardo Coello1, Nicolas R. Bolo2, Huijun Liao1, Angelina Awad3, Margaret A. Niznikiewicz3, and Alexander Lin1
1Radiology, Brigham and Women's Hospital, Boston, MA, United States, 2Psychiatry, Beth Israel Deaconess Medical Center, Boston, MA, United States, 3Psychiatry, Veterans Affairs Boston Healthcare System, Boston, MA, United States
This work describes a methodology to perform reproducible functional MR spectroscopy (FMRS) experiments to study auditory processing at 7T. A processing pipeline and quality control steps are proposed to remove distortions and temporal instabilities in the dynamic experiment. The designed experiment was successfully tested, producing highly correlated results in healthy volunteers and patients with schizophrenia scanned multiple times.
|
|
2917.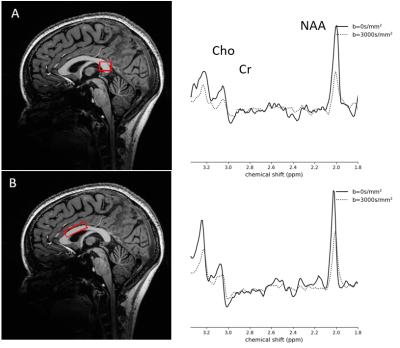 |
Diffusion MR spectroscopy at 7T with no water suppression is sensitive to measuring diffusion properties of metabolites in the corpus callosum
Yasmin Mzayek1,2, Francesca Branzoli3, Thomas Troalen4, Yann Le Fur1,2, Patrick Viout1,2, Tangi Roussel1,2, Stanislas Rapacchi1,2, Maxime Guye1,2, Itamar Ronen5, Wafaa ZAARAOUI1,2, and Jean-Philippe RANJEVA1,2
1Aix-Marseille Université, CNRS, CRMBM, Marseille, France, 2APHM, Hôpital de la Timone, Pôle d’Imagerie Médicale, CEMEREM, Marseille, France, 3Brain and Spine Institute - ICM, Centre for NeuroImaging Research - CENIR, Paris, France, 4Siemens Healthineers SAS, Saint-Denis, France, 5Department of Radiology Leiden University Medical Center, Leiden, Netherlands
We implemented a diffusion-weighted semi-LASER single voxel spectroscopy sequence with no water suppression to measure mean diffusivity and fractional anisotropy within two volumes-of-interest in the corpus callosum. Without water suppression, we were able to decrease acquisition time and use the water signal for phase, frequency, and eddy current corrections. We measured the signal decay of N-acetylaspartate (tNAA), creatine (tCr), choline (tCho), and water to derive diffusion outcomes and compare between the two volumes-of-interest. Our implementation was done in feasible time for in vivo measurements. Further development can lead to more specific assessments of brain microstructure and pathophysiology.
|
|
2918.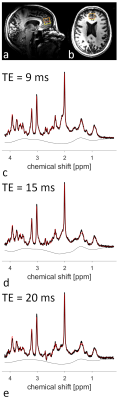 |
Determination of Metabolite T2-Relaxation in the Medial Prefrontal Cortex at 7T Using an 8Tx/8Rx Channel Head Coil
Ariane Fillmer1, Layla Tabea Riemann1, Frank Seifert1, Semiha Aydin1, Harald Pfeiffer1, and Bernd Ittermann1
1Physikalisch-Technische Bundesanstalt (PTB), Braunschweig und Berlin, Germany
To investigate underlying mechanisms of psychiatric diseases, such as schizophrenia, magnetic resonance spectroscopy is a powerful tool. Here, the medial prefrontal cortex is a region of high interest, however, high quality measurements are technically challenging, and proper absolute quantification is hindered by the lack of literature values for metabolite relaxation times. This work presents the acquisition of high-quality MR spectra from the medial prefrontal cortex, and the preliminary calculation of metabolite T2 relaxation times, using an 8Tx/8Rx channel coil with a dedicated phase set optimized for B1 efficiency in the frontal cortex.
|
|
2919.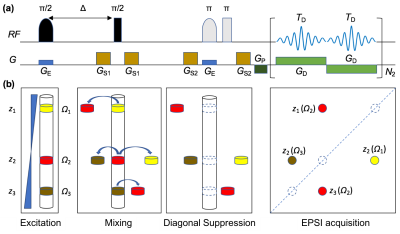 |
Ultrafast diagonal-suppressed 2D MR correlation spectroscopy
Liangjie Lin1,2 and Zhong Chen2
1Philips Healthcare, Beijing, China, 2Department of Electronic Science, Xiamen University, Xiamen, China
Two-dimensional (2D) MR spectroscopy enables feasible detection of low-concentration scalar-coupled metabolites in human tissues. However, the traditional acquisition of 2D MRS can be time consuming, and cross peaks which contain crucial spectral information in 2D correlated spectra (COSY) usually get contaminated by nearby strong diagonal peaks. Here, a pulse sequence is developed for ultrafast recording of diagonal-free 2D MR correlated spectra, thus allowing improved recognition and measurement of scalar-coupled metabolites.
|
|
2920.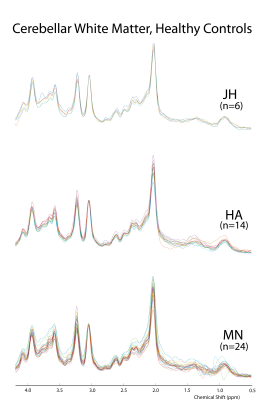 |
Multi-site/Multi-vendor reproducibility of advanced MRS at 3T in a clinical cohort
James M Joers1, Dinesh K Deelchand1, Bin Guo2, Lynn E Eberly2, Young Woo Park1, Adam Berrington3, Michal Povazan4, Andre van der Kouwe5, Khalaf Bushara6, Chiadi U Onyike7, Jeremy Schmahmann8, Peter B Barker4,9, Eva Ratai5,
and Gülin Öz1
1CMRR/Radiology, University of Minnesota, Minneapolis, MN, United States, 2Division of Biostatistics, University of Minnesota, Minneapolis, MN, United States, 3Sir Peter Mansfield Imaging Centre, School of Physics and Astronomy, University of Nottingham, Nottingham, United Kingdom, 4Radiology and Radiological Science, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 5A.A. Martinos Center for Biomedical Imaging / Radiology, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States, 6Neurology, University of Minnesota Medical School, Minneapolis, MN, United States, 7Psychiatry and Behavioral Sciences, Johns Hopkins University, Baltimore, MD, United States, 8Neurology, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States, 9Kennedy Krieger Institute, Johns Hopkins University School of Medicine, Baltimore, MD, United States
The reproducibility of an advanced single voxel MRS protocol was tested in a clinical cohort and control subjects at 3T in a multi-site setting. A standardized MRS protocol was implemented on two MR system vendors (Siemens, Philips) over three sites. This protocol utilized automated voxel placement and an sLASER pulse sequence with identical radiofrequency waveforms, pulse durations, interpulse delays and gradient spoilers. Spectral quality, metabolite concentrations and test-retest reproducibility results from three voxels are reported from both the healthy control group and subjects with genetically-confirmed spinocerebellar ataxia type 3 (SCA3), a hereditary movement disorder.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Video
Watch the Video View the Poster
View the Poster Back to Top
Back to Top