Digital Poster Session
Acquisition, Reconstruction & Analysis: Novel Acquisition, Reconstruction, and Analysis
Acquisition, Reconstruction & Analysis
3430 -3444 Novel Acquisition, Reconstruction, and Analysis - Novel Acquisitions & Reconstructions: Parallel Imaging
3445 -3460 Novel Acquisition, Reconstruction, and Analysis - Novel Acquisitions & Reconstructions: Sparse & Low-Rank
3461 -3476 Novel Acquisition, Reconstruction, and Analysis - Novel Acquisitions & Reconstructions: Other
3477 -3492 Novel Acquisition, Reconstruction, and Analysis - Acquisition, Reconstruction & Analysis 1
3493 -3507 Novel Acquisition, Reconstruction, and Analysis - Acquisition, Reconstruction & Analysis 2
3430.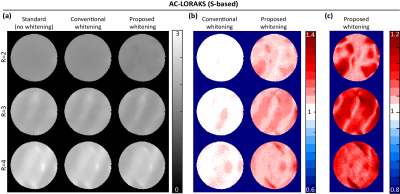 |
On Coil Combination with Optimal SNR for Linear Multichannel k-Space Reconstruction Methods
Daeun Kim1, Jonathan Polimeni2, Kawin Setsompop2, and Justin Haldar1
1Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Harvard Medical School, Massachusetts General Hospital, Charlestown, MA, United States
Noise correlations exist in multi-channel k-space data, and methods to optimally account for this correlation have been used for a long time in image-domain parallel imaging methods like SENSE. However, methods to address noise are not widely-utilized for Fourier-domain parallel imaging methods like GRAPPA, SPIRiT, and AC-LORAKS. In this work, we demonstrate that properly accounting for spatially-varying noise correlation can substantially reduce the noise level of coil-combined images. Further, we demonstrate the existence of previously-unknown correlations between the real and imaginary parts of the noise in reconstructed images. Accounting for this extra correlation can reduce the noise level even further.
|
|
3431.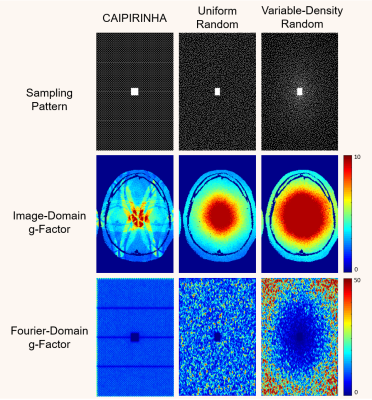 |
Transform-Domain g-Factor Maps
Jiayang Wang1 and Justin P. Haldar1
1Department of Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States
The g-factor is commonly used for quantifying the noise amplification associated with accelerated data acquisition and linear image reconstruction, and is frequently used to compare different k-space sampling strategies. While previous work computes g-factors in the image domain, we observe in this work that g-factors can also be used to quantify uncertainty in various transform domains (e.g., the wavelet domain and the multi-channel Fourier domain). These transform-domain g-factor maps provide complementary information to conventional image-domain g-factor maps, and are potentially useful for k-space sampling pattern design.
|
|
3432.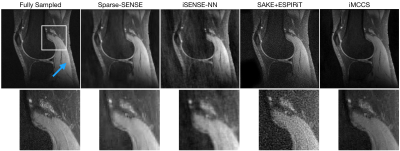 |
Compressed Sensing Parallel Imaging Without Calibration
Nicholas Dwork1, Corey A. Baron2, Ethan M. I. Johnson3, Daniel O'Connor4, Adam B. Kerr5, Peder E. Z. Larson1, Jeremy Gordon1, and John M. Pauly6
1Radiology and Biomedical Imaging, University of California in San Francisco, San Francisco, CA, United States, 2Medical Biophysics, Western University, London, ON, Canada, 3Biomedical Engineering, Northwestern University, Evanston, IL, United States, 4Mathematics and Statistics, University of San Francisco, San Francisco, CA, United States, 5Center for Cognitive and Neurobiological Imaging, Stanford University, Stanford, CA, United States, 6Electrical Engineering, Stanford University, Stanford, CA, United States In this paper, the reconstructed image is the result of a compressed sensing optimization problem that includes constraints based on fundamental physics. The problem is solved using an alternating minimization approach: two convex optimization problems are alternately solved, one with the Fast Iterative Shrinkage Threshold Algorithm (FISTA) and the other with the Primal-Dual Hybrid Gradient method. Results show improved detail when compared to conventional SENSE results. |
|
3433.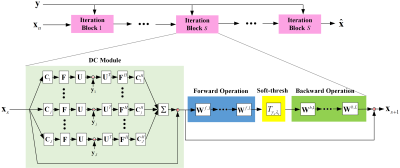 |
pISTA-SENSE-ResNet for Parallel MRI Reconstruction
Tieyuan Lu1, Xinlin Zhang1, Yihui Huang1, Di Guo2, and Xiaobo Qu1
1Department of Electronic Science, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Xiamen University of Technology, Xiamen, China
Magnetic resonance imaging has been widely applied in clinical diagnosis, however, is limited by its long data acquisition time. Although imaging can be accelerated by sparse sampling and parallel imaging, achieving promising reconstruction images with a fast reconstruction speed remains a challenge. In this work, we design the neural network structure from the perspective of sparse iterative reconstruction. The experimental results of a public knee dataset show that compared with the optimization-based method and the latest deep learning parallel imaging methods, the proposed network has less error in reconstruction and is more stable under different acceleration factors.
|
|
3434.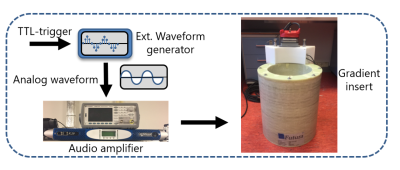 |
Accelerating silent MRI using a gradient axis at 20 kHz and generalized conjugate gradient SENSE reconstruction
Edwin Versteeg1, Tijl Van der Velden1, Jeroen Hendrikse1, Dennis Klomp1, and Jeroen Siero1,2
1Radiology, University Medical Center Utrecht, Utrecht, Netherlands, 2Spinoza Centre for Neuroimaging Amsterdam, Amsterdam, Netherlands
A silent gradient axis driven at 20 kHz can be used to decrease acoustic noise and increase patient comfort in MR-exams. However, the speed of this silent acquisition is determined by the amount of data needed for artifact free reconstructions. In this work, we present a framework for increasing the time efficiency while maintaining image quality of silent imaging with a silent gradient axis. We show that, by using a generalized conjugate gradient SENSE reconstruction, a 2-3-fold decrease in scan time is feasible both on a phantom and in-vivo.
|
|
3435.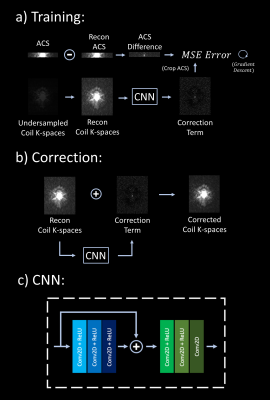 |
Scan-specific, Parameter-free Artifact Reduction in K-space (SPARK)
Onur Beker1,2, Congyu Liao1,3, Jaejin Cho1,3, Zijing Zhang1,4, Kawin Setsompop1,3,5, and Berkin Bilgic1,3,5
1Martinos Center for Biomedical Imaging, Charlestown, MA, United States, 2Electrical and Electronics Engineering, Bogazici University, Istanbul, Turkey, 3Harvard Medical School, Boston, MA, United States, 4College of Optical Science and Engineering, Zhejiang University, Hangzhou, China, 5Harvard/MIT Health Sciences and Technology, Cambridge, MA, United States
We propose a convolutional neural network (CNN) approach that works synergistically with physics-based reconstruction methods to reduce artifacts in accelerated MRI. Given reconstructed coil k-spaces, our network predicts a k-space correction term for each coil. This is done by matching the difference between the acquired autocalibration lines and their erroneous reconstructions, and generalizing this error term over the entire k-space. Application of this approach on existing reconstruction methods show that SPARK suppresses reconstruction artifacts at high acceleration, while preserving and improving on detail in moderate acceleration rates where existing reconstruction algorithms already perform well; indicating robustness.
|
|
3436. |
Model based Deep Learning for Calibrationless Parallel MRI recovery
Aniket Pramanik1, Hemant Kumar Aggarwal1, and Mathews Jacob1
1Electrical and Computer Engineering, The University of Iowa, Iowa City, IA, United States
We introduce a fast model based deep learning approach for calibrationless parallel MRI reconstruction. The proposed scheme is a non-linear generalization of structured low-rank (SLR) methods that self learn linear annihilation filters. It pre-learns non-linear annihilation relations in the Fourier domain from exemplar data which significantly reduces the computational complexity, making it three orders of magnitude faster than SLR schemes. It allows incorporation of spatial domain prior that offers improved performance over calibrated image domain MoDL approach. The calibrationless strategy minimizes potential mismatches between calibration data and the main scan, while eliminating the need for a fully sampled calibration region.
|
|
3437.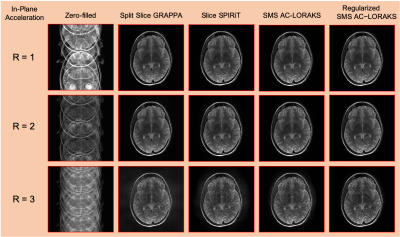 |
Advanced New Linear Predictive Reconstruction Methods for Simultaneous Multislice Imaging
Rodrigo A. Lobos1, Tae Hyung Kim1, Kawin Setsompop2, and Justin P. Haldar1
1Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Martinos Center for Biomedical Imaging, Charlestown, MA, United States
Many autocalibrated parallel imaging reconstruction methods are based on linear-predictive/autoregressive principles, including noniterative GRAPPA-type interpolation methods, iterative SPIRiT-type annihilation methods, and structured low-rank matrix methods like PRUNO and Autocalibrated LORAKS. In principle, all of these approaches could be adapted for simultaneous multislice (SMS) reconstruction. However, in practice, GRAPPA-type SMS methods are popular, but there has been limited exploration of more advanced annihilation-based or structured low-rank matrix SMS methods. In this work, we adapt and evaluate these advanced approaches for SMS reconstruction. Results demonstrate that these advanced approaches can offer substantial improvements over simpler GRAPPA-type methods when applied to SMS.
|
|
3438.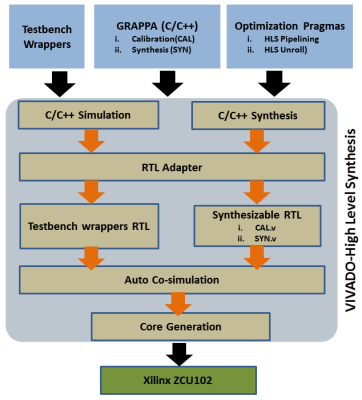 |
FPGA-based Coprocessor for Real-time GRAPPA Reconstruction (GR-co-RECON)
Abdul Basit1,2, Omair Inam1, Mahmood Qureshi1, and Hammad Omer1
1Electrical and Computer Engineering, Comsats University Islamabad, islamabad, Pakistan, 2Computer Engineering, Khwaja Fareed University of Engineering and Information Technology, Rahim Yar Khan, Pakistan
Data acquisition and reconstruction speed both are crucial for real-time MRI. However, MR image reconstruction speed is highly dependent on the processing capabilities of the hardware platforms (e.g. CPUs, GPUs). Recently, it has been observed that Field Programmable Gate Arrays (FPGA) are a potential candidate to address the computational demands of parallel MRI algorithms. This paper presents the first design effort to implement high performance 32-bit floating-point FPGA-based coprocessor for real-time GRAPPA reconstruction. In-vivo results of 12-channel, 3.0T human-head dataset show that the proposed system speeds up the image reconstruction time up to 106x without compromising image quality.
|
|
3439.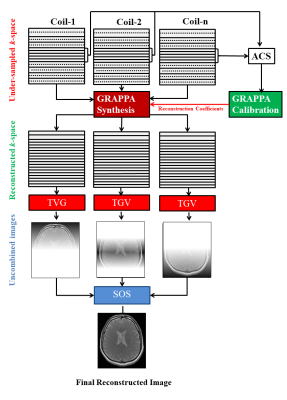 |
MR Image Reconstruction using GRAPPA with Total Generalized Variation
Rida Zainab1, Muhammad Haseeb Hassan1, Omair Inam1, Ibtisam Aslam1,2, and Hammad Omer1
1Department of Electrical and Computer Engineering, COMSATS University Islamabad, Islamabad, Pakistan, 2Department of Radiology and Medical Informatics, Hospital University of Geneva, Geneva, Switzerland
GRAPPA reconstructed images may exhibit noise modulated by the receiver coil sensitivities. Total variation (TV) regularization has been recently used to solve the image de-noising problem. However, conventional TV fails to remove staircase artifacts in the reconstructed MR images due to inhomogeneities in field strength and receiver coils. In this abstract, total generalized variation (TGV) regularization is used to de-noise the GRAPPA reconstructed images, while eliminating the limitations posed by TV. Experiments are performed on 8-channel in-vivo human-head data set. The results show that the proposed method successfully removes the noise and preserve fine details in the GRAPPA reconstructed images.
|
|
3440.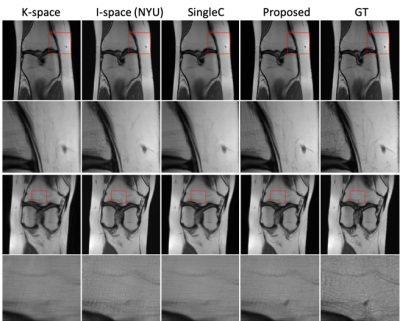 |
Parallel MRI Reconstruction via Residual UNet with Joint Consideration of k-Space and Image Space
Xiaoxia Zhang1, Xiaopeng Zong1, Yong Chen1, Zhenghan Fang1, and Pew-Thian Yap1
1Department of Radiology, Department of Radiology and Biomedical Research Imaging Center (BRIC), University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
We propose a strategy based on residual U-Net to reconstruct MR images from undersampled multichannel data by considering both k-space and image space. Our method first imputes the missing data points in k-space by utilizing the intrinsic relationships among channels. Then, the image reconstructed from the imputed k-space data is fed to another network for spatial detail refinement. Our method does not necessarily require auto-calibration signal (ACS) and is hence less susceptible to motion-induced inconsistency between the ACS and the undersampled data. Comprehensive evaluation indicates that our method yields images with superior perceptual details.
|
|
3441.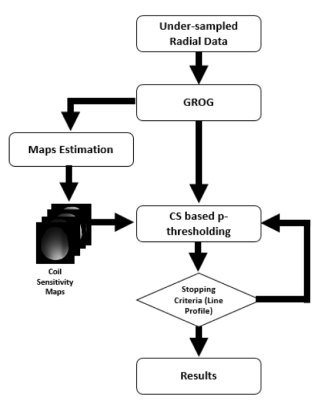 |
Calibration-less pMRI for the Reconstruction of Radially Encoded data using GROG based CS
Husnain Javid Bhatti1,2, Fariha Aamir1, Ibtisam Aslam1,3, Khan Afsar1,4, and Hammad Omer1
1Department of Electrical and Computer Engineering, COMSATS University Islamabad (CUI), Islamabad, Pakistan, 2School of Electrical Engineering and Computers Science, National University of Science and Technology (NUST), Islamabad, Pakistan, 3Department of Radiology and Medical Informatics, Hospital University of Geneva, GENEVA, Switzerland, 4Department of Electronic science and Technology, Xiamen University, Xiamen, China
To reduce MRI scan time, under-sampled non-Cartesian trajectories are used which lead to artifacts. This work proposes a new method ‘GROG with calibration-less pMRI for CS based p-thresholding’ to reconstruct MR images from the under-sampled radial k-space data. The proposed method is validated on the phantom and 1.5T human head data and provides significant improvement both visually and in terms of quantifying parameters (AP, RMSE & PSNR) e.g. 77% and 86% improvement in AP, 5% and 32% improvement in RMSE, 7% and 11% improvement in PSNR at AF=4 for the phantom data than POCS and pseudo Cartesian GRAPPA, respectively.
|
|
3442. |
High-frequency Transform Guided Denoising Autoencoding Prior for Parallel MR Imaging
Yuanyuan Hu1, Zhuonan He2, Jinjie Zhou2, Minghui Zhang2, Qiegen Liu2, and Dong Liang1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518055, China, Shenzhen, China, 2the Department of Electronic Information Engineering, Nanchang University, Nanchang 330031, China, Nanchang, China
Ill-posed inverse problems embodied in parallel imaging remain an active research topic in several decades, with new approaches constantly emerging. Built on the observation that both dictionary learning and conventional sparse coding extract high-frequency component to model, we derived a novel strategy named HDAEP to explore the prior on high-frequency domain on the basis of denoising autoencoding. After the prior is learned from the trained network, the iteratively Gauss-Newton method is employed to jointly estimating the images and coil sensitivities. Experimental results show that the proposed method can achieve superior performances on parallel MRI reconstruction compared to state-of-the-arts.
|
|
3443.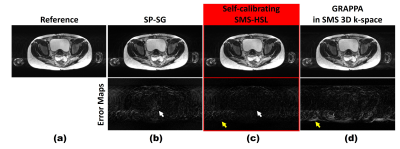 |
Robust Simultaneous Multi-Slice MRI Exploiting Hankel Subspace Learning with Self-Calibration and Self-Referencing Magnitude Prior
Eun Ji Lim1, Guobin Li2, Chaohong Wang2, Zhaopeng Li2, Shasha Yang2, and Jaeseok Park1
1Sungkyunkwan University, Suwon, Republic of Korea, 2United Imaging Healthcare, Shanghai, China
SMS methods typically consist of the following two steps: kernel calibration and reconstruction. However, discrepancies between calibration and imaging occur due to either different image contrasts or subject motions, resulting in residual aliasing artifacts. To tackle these problems, in this work we propose a robust SMS technique exploiting Hankel subspace learning with self-calibration and self-referencing magnitude prior. An SMS filter is designed to strictly control pass-bands and stop-bands to reduce the dependence of image contrast on reconstruction. Both external and internal calibrating signals are included in the calibration step, while a self-referencing magnitude prior is imposed in the reconstruction step.
|
|
3444.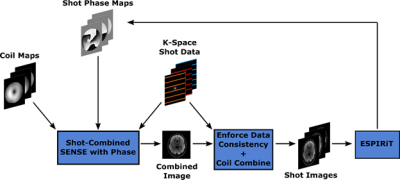 |
Phase Reconstruction using Iterative Multi-shot ESPIRiT (PRIME)
Stephen Cauley1,2, Bryan Clifford3, Steffen Bollmann3, Thorsten Feiweier4, Berkin Bilgic1,2, Kawin Setsompop1,2,5, and Lawrence L. Wald1,2,5
1Department of Radiology, A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 2Harvard Medical School, Boston, MA, United States, 3Siemens Medical Solutions USA, Boston, MA, United States, 4Siemens Healthcare GmbH, Erlangen, Germany, 5Harvard-MIT Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
We employ a compact phase modeling strategy for accurate multi-shot echo-planar imaging (msEPI) reconstruction. As an alternative to approaches that perform msEPI reconstruction using strict low-rank constraints, we recast the problem as an iterative relative phase estimation problem. This allows for us to utilize existing techniques such as ESPIRiT, which are formulated for determining relative magnitude and phase differences between multi-coil receive arrays. Through an iterative search we jointly estimate an artifact-free combined image and the smooth relative phase between each msEPI shot. We demonstrate the benefits of our approach for clinical and highly-accelerated multi-shot diffusion-weighted acquisitions.
|
3445.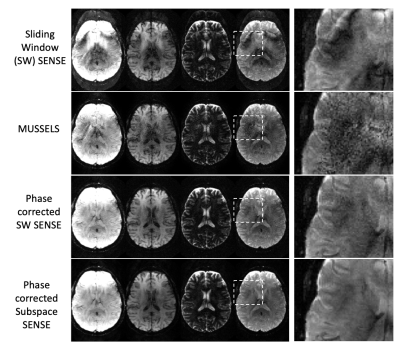 |
A rapid quantitative Multi-inversion SAGE-EPI brain protocol with subspace reconstruction and navigation-free shot-to-shot phase correction
Mary Kate Manhard1,2, Zijing Dong1,3, Congyu Liao1,2, Merlin Fair1,2, Fuyixue Wang1,4, Berkin Bilgic1,2, and Kawin Setsompop1,2,4
1A.A. Martinos Center for Biomedical Imaging, Boston, MA, United States, 2Department of Radiology, Harvard Medical School, Boston, MA, United States, 3Department of Electrical Engineering and Computer Science, Massachusetts Institution of Technology, Cambridge, MA, United States, 4Harvard-MIT Division of Health Sciences and Technology, Cambridge, MA, United States
Recently, multi-contrast EPI approaches have been proposed for a fast screening brain protocol. With EPI-encoding, multiple contrasts can be acquired quickly for robust, quantitative mapping as well as creation of synthetic weighted images. Here, a spatiotemporal subspace reconstruction is developed to jointly reconstruct multi-contrast multishot-EPI data from a multi-inversion Spin and Gradient echo EPI (MI-SAGE-EPI) acquisition. An approach to estimate and incorporate shot-to-shot phase corruption into the reconstruction was also developed. This navigation-free subspace reconstruction achieves good reconstruction for MI-SAGE-EPI at a high EPI-acceleration, thus enabling a rapid quantitative protocol at high in-plane resolution with minimal distortion and blurring.
|
|
3446.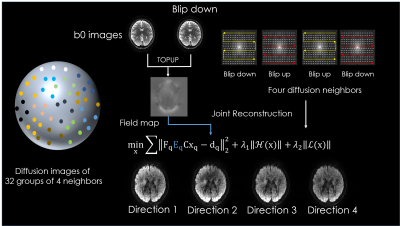 |
Highly accelerated distortion free diffusion imaging using joint k/q-space reconstruction.
Wonil Lee1, Seohee So1, Jaejein Cho2, Congyu Liao2, Qiyuan Tian2, Hyunwook Park1, Elfar Adalsteinsson3, Kawin Setsompop4, and Berkin Bilgic2
1Electrical engineering, KAIST, Daejeon, Korea, Republic of, 2Martinos Center, Charlestown, MA, United States, 3MIT, Cambridge, MA, United States, 4Martinos Center, Charlestown, China
We propose k/q Blip-Up and -Down Acquisition (k/q-BUDA) for distortion-free diffusion MRI (dMRI). We acquire different diffusion directions with alternating phase-encoding polarities, and incorporate $$$B_{0}$$$ information in the forward model of our reconstruction. Multiple images on neighboring q-space points are jointly reconstructed using Hankel structured low-rank regularization in k-space. This allows us to exploit similarities across q-space at the parallel imaging level, thus enabling k/q-space joint reconstruction. We select four neighboring q-space samples, providing us with two blip-up and two blip-down phase-encoded shots, who provide complementary $$$B_{0}$$$ encoding to eliminate distortion in the reconstructed volumes.
|
|
3447.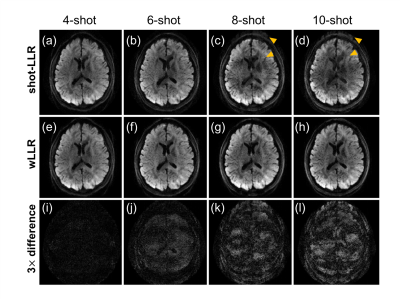 |
Weighted-nuclear-norm minimization for low-rank approximation -- application to multi-shot diffusion-weighted MRI reconstruction
Yuxin Hu1,2, Qiyuan Tian1,2, Yunyingying Xu2, Philip K. Lee1,2, Bruce L. Daniel1,3, and Brian A. Hargreaves1,2,3
1Department of Radiology, Stanford University, Stanford, CA, United States, 2Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 3Department of Bioengineering, Stanford University, Stanford, CA, United States
Low rankness of image/k-space data has been exploited in many different MRI applications. In this work, we introduce weighted-nuclear-norm minimization for MRI low-rank reconstruction, in which smaller thresholds are used for larger eigenvalues to reduce information loss. Our simulation results demonstrate that weighted nuclear norm could serve as a better rank approximation compared to (unweighted) nuclear norm. With this technique, we achieve 10-shot DWI and high-fidelity half-millimetre DWI reconstruction with significantly reduced ghosting artifacts.
|
|
3448.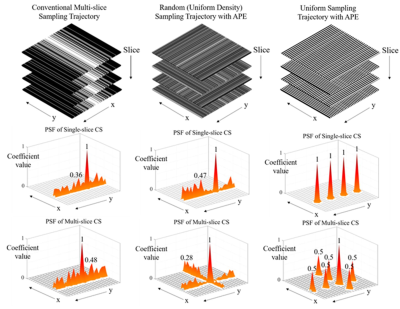 |
A Novel Compressed Sensing Multi-Slice Cartesian MRI via Alternating Phase Encoding Directions for Acceleration and Flexible Undersampling
Zheyuan Yi1,2,3, Yilong Liu1,2, Fei Chen3, and Ed X. Wu1,2
1Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 2Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China
In conventional compressed sensing (CS) multi-slice Cartesian 2D imaging, the undersampling is performed along phase-encoding direction only, leading to coherent 1D aliasing that significantly limits the effectivess of CS for acceleration. This study proposes a multi-slice CS reconstruction method to take advantage of extremely augmented sampling incoherence created by orthogonally alternating phase-encoding directions among adjacent slices. The multi-slice CS approach was evaluated with single-channel brain T1W and T2W datasets. The results demonstrate significant improvements with both pseudo-random and uniform undersampling. This new method will also greatly augment the existing 2D CS parallel imaging technqiues for very high acceleration.
|
|
3449.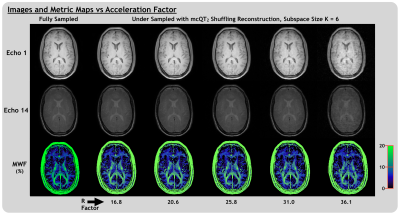 |
Multi-Component Quantitative T2 Shuffling: Accelerating Myelin Water Imaging by 20-30x
Adam V. Dvorak1,2, Guillaume Gilbert3, Alex L. MacKay1,4, and Shannon H. Kolind1,2,4,5
1Physics and Astronomy, University of British Columbia, Vancouver, BC, Canada, 2International Collaboration on Repair Discoveries (ICORD), Vancouver, BC, Canada, 3MR Clinical Science, Philips Canada, Markham, ON, Canada, 4Radiology, University of British Columbia, Vancouver, BC, Canada, 5Medicine (Neurology), University of British Columbia, Vancouver, BC, Canada
Multi-component quantitative T2 mapping can provide a range of valuable, in-vivo biomarkers, but is limited by lengthy acquisition times. Here we introduce multi-component quantitative T2 shuffling, a subspace-constrained CS method for reconstructing highly under-sampled multi-component relaxation mapping data using principal components of a temporal basis. We demonstrated reconstruction of myelin water imaging data with simulated under-sampling acceleration factors of ~20-30, which could provide accurate images, higher resolution and fit-to-noise ratios, and improved metric maps in a fraction of the acquisition time.
|
|
3450.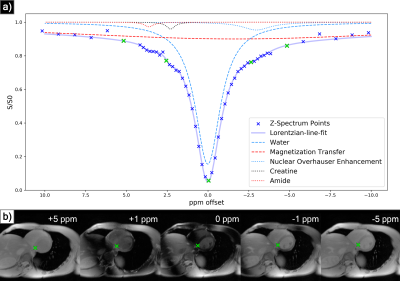 |
Multiscale Low Rank Matrix Decomposition for Reconstruction of Accelerated Cardiac CEST MRI
Ilya Chugunov1, Wissam AlGhuraibawi2, Kevin Godines2, Bonnie Lam2, Frank Ong3, Jonathan Tamir1,4, and Moriel Vandsburger2
1Electrical Engineering and Computer Science, University of California, Berkeley, Berkeley, CA, United States, 2Bioengineering, University of California, Berkeley, Berkeley, CA, United States, 3Electrical Engineering, Stanford University, Stanford, CA, United States, 4Electrical and Computer Engineering, University of Texas at Austin, Austin, TX, United States
Multiscale low rank reconstruction has been demonstrated to efficiently reconstruct non-gated dynamic MRI by leveraging sparsity in the time domain. This abstract demonstrates its ability to reconstruct 4-fold accelerated CEST imaging of the heart via similarly exploiting sparsity in the Z-spectrum domain. This reconstruction outperforms zero-filled IFFT for quantization of magnetization transfer, nuclear overhauser, and CEST effects as derived from Lorentzian-line-fit analysis. Extension to volumetric or motion inclusive CEST imaging and development of a new regularization function may enable further acceleration.
|
|
3451.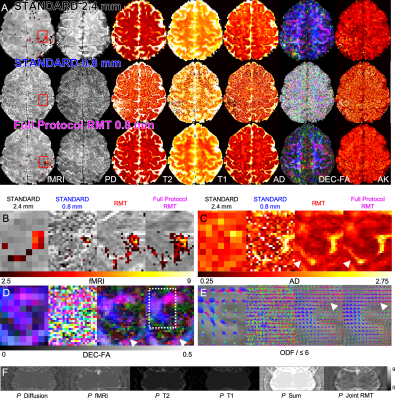 |
MRI Below the Noise Floor
Gregory Lemberskiy1,2, Steven Baete1, Jelle Veraart1, Timothy M. Shepherd1, Els Fieremans1, and Dmitry S Novikov1
1New York University School of Medicine, New York, NY, United States, 2Microstructure Imaging INC, New York, NY, United States
We develop random matrix theory (RMT)-based MRI image reconstruction able to increase SNR by up to 10-fold, and to radically increase resolution for routine clinical acquisitions. RMT offers an objective criterion for separating signal from noise across all coils, voxels and MRI contrasts, by utilizing the redundancy in MRI measurements. We demonstrate RMT on a 0.8x0.8x0.8 mm3 neuro exam that includes a series of multiple T2w, T1w, diffusion, and fMRI images on a 3T clinical scanner. RMT can serve as a paradigm for reconstructing multiple contrasts, enhancing image quality for low-field scanners, increasing MR value, and improving biomarker precision.
|
|
3452.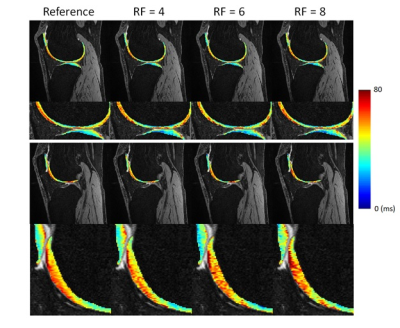 |
Highly accelerated T1ρ imaging with Kernel-based low-rank compressed sensing: evaluation of retrospective and prospective undersampling
Jeehun Kim1,2, Chaoyi Zhang3, Mingrui Yang1,2, Hongyu Li3, Mei Li1,2, Richard Lartey1,2, Leslie Ying3,4, and Xiaojuan Li1,2,5
1Biomedical Engineering, Cleveland Clinic, Cleveland, OH, United States, 2Program of Advanced Musculoskeletal Imaging (PAMI), Cleveland Clinic, Cleveland, OH, United States, 3Electrical Engineering, University at Buffalo, the State University of New York, Buffalo, NY, United States, 4Biomedical Engineering, University at Buffalo, the State University of New York, Buffalo, NY, United States, 5Department of Diagnostic Radiology, Cleveland Clinic, Cleveland, OH, United States
Quantitative T1ρ MRI provides valuable information on compositional changes in cartilage, but requires longer scan time compared to conventional imaging. In this work, kernel-based low-rank compressed sensing reconstruction was used to accelerate T1ρ imaging and the retrospective and prospective undersampled results from four subjects were compared to reference T1ρ values.
|
|
3453.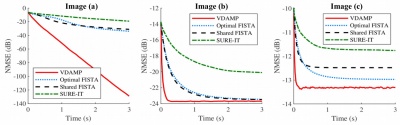 |
Versatile Parameter-Free Compressed Sensing MRI with Approximate Message Passing
Charles Millard1,2, Aaron T Hess2, Boris Mailhé3, and Jared Tanner1
1Mathematical Institute, University of Oxford, Oxford, United Kingdom, 2Oxford Centre for Clinical Magnetic Resonance, University of Oxford, Oxford, United Kingdom, 3Siemens Healthineers, Princeton, NJ, United States
We consider two algorithmic challenges for the compressed sensing MRI community: (1) the difficulty of tuning free model parameters and (2) the need to converge quickly. The authors have developed a parameter-free approach to reconstruction which accommodates structurally rich regularizers that can be automatically adapted to near-optimality, removing the need for manual adjustment between images or sampling schemes. We evaluate the algorithm’s performance on three test images of varying type and dimension and find that it converges faster and to a lower mean-squared error than its competitors, even when they are optimally tuned.
|
|
3454.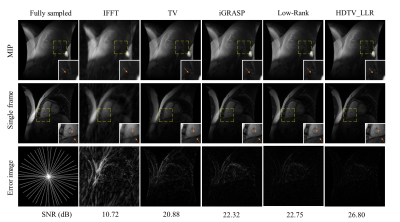 |
Accelerated 4D MRI using joint higher degree total variation and local low-rank constraints (HDTV-LLR)
Yue Hu1, Disi Lin1, and Dong Nan2
1Harbin Institute of Technology, Harbin, China, 2The First Affiliated Hospital of Harbin Medical University, Harbin, China
Four-dimensional (4D) MRI can provide 3D tissue properties and the temporal profiles at the same time. However, further applications of 4D MRI is limited by the long acquisition time and motion artifacts. We propose a new 4D MRI reconstruction scheme, named HDTV-LLR, by integrating the three-dimensional higher degree total variation and the local low-rank penalties. We demonstrate the benefits of the proposed method using 4D cardiac MRI dataset with undersampling factors of 12 and 16. The proposed method is compared with iGRASP, and schemes using either low-rank or sparsity constraint alone. Results show improved image quality and reduced artifacts.
|
|
3455.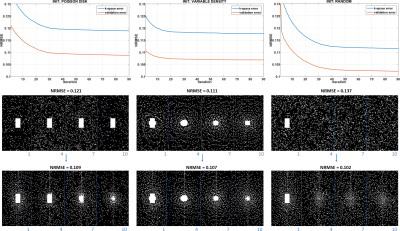 |
Learning-Based Sampling Pattern for Compressed Sensing and Low Rank Reconstructions using Multicoil MR Images of Human Knee Joint
Marcelo V. W. Zibetti1, Gabor T. Herman2, and Ravinder R. Regatte1
1Radiology, NYU, New York, NY, United States, 2The Graduate Center, CUNY, New York, NY, United States
Compressed Sensing (CS) and parallel MRI (pMRI) have been successfully applied to accelerate MRI-data acquisition. CS requires incoherence, usually achieved by random undersampling the data, but pMRI does not. Combined, these methods allow even higher acceleration rates. However, it is unknown how the sampling pattern (SP) should be selected. It is also unknown if the SP is dependent on the reconstruction method. Here we demonstrate, using a new algorithm, that the SP can be learned from given data and reconstruction method. Our results show that the learned SP is superior to others such as Poisson disk and variable density.
|
|
3456.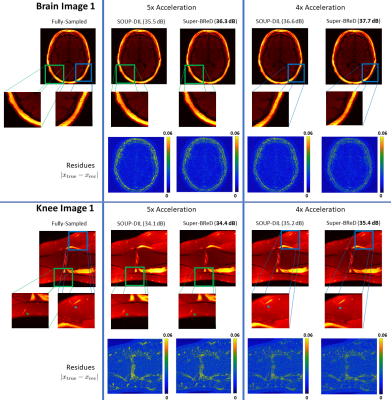 |
Combining Supervised and semi-Blind Dictionary (Super-BReD) Learning for MRI Reconstruction
Anish Lahiri1, Saiprasad Ravishankar2, and Jeffrey A Fessler1
1Electrical and Computer Engineering, University of Michigan, Ann Arbor, MI, United States, 2Computational Mathematics, Science and Engineering, and Biomedical Engineering, Michigan State University, East Lansing, MI, United States
Regularization in MRI reconstruction often involves sparse representation of signals using linear combinations of dictionary atoms. In 'blind' settings, these dictionaries are learned during reconstruction from the corrupt/aliased images, using no training data. In contrast, 'Fully supervised' dictionary learning (DL) requires uncorrupted/fully sampled training images, and the learned dictionary is used to regularize image reconstruction from undersampled data. We combine the aforementioned DL frameworks to learn two separate dictionaries in a residual fashion to jointly reconstruct an undersampled image. Our algorithm, Super-BReD Learning, shows promising results on reconstruction from retrospectively undersampled data, and outperforms recent DL schemes.
|
|
3457.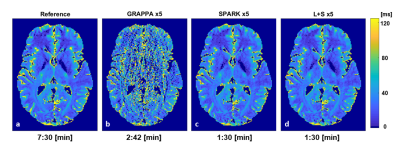 |
Comparison of fixed vs. low rank reconstruction methods for acceleration of quantitative T2 mapping
Daniel Grzeda1, Meirav Galun2, Noam Omer1, Tamar Blumenfeld-Katzir1, Dvir Radunsky1, Ricardo Otazo3, and Noam Ben-Eliezer1,4,5
1Department of Biomedical Engineering, Tel Aviv University, Tel Aviv, Israel, 2Department of Computer Science and Applied Mathematics, Weizmann Institute of Science, Rehovot, Israel, 3Departments of Medical Physics and Radiology, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 4Center for Advanced Imaging Innovation and Research (CAI2R), New-York University Langone Medical Center, New York, New York, NY, United States, 5Sagol School of Neuroscience, Tel-Aviv University, Tel Aviv, Israel
Quantification of T2 values is valuable for a wide range of research and clinical applications. Multi-echo spin echo protocols allow mapping T2 values, yet, at the cost of strong contamination from stimulated echoes. The echo-modulation-curve algorithm can efficiently overcome these limitations to produce accurate T2 values. Still, integration into clinical routine requires further acceleration in scan time. In this work we present a fixed-rank and sparse algorithm (SPARK) for accelerating the acquisition of T2 values, and compare it against standard L+S and GRAPPA. SPARK was found to improve robustness to reconstruction parameters, and achieve superior accuracy at high acceleration factors.
|
|
3458.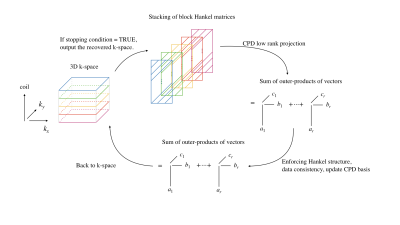 |
Low-Rank Tensor Completion for Accelerated Magnetic Resonance Imaging
Shen Zhao1, Lee C. Potter1, and Rizwan Ahmad2
1Department of Electrical and Computer Engineering, The Ohio State University, Columbus, OH, United States, 2Department of Biomedical Engineering, The Ohio State University, Columbus, OH, United States
We present a method for calibration-less, accelerated Magnetic Resonance Imaging (MRI) via canonical polyadic decomposition (CPD) based low-rank tensor completion (LRTC). LRTC exploits the higher dimensional structure inherent in MRI. Preliminary results show that LRTC can outperform structured low-rank matrix completion methods for 2D and compressed sensing-based methods for dynamic applications.
|
|
3459.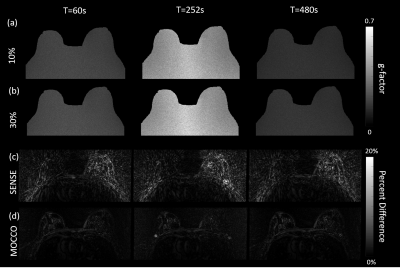 |
Data-Driven Model Consistency Condition Reconstruction Performance for Radial Acquisition in Breast DCE-MRI
Ping N Wang1, Julia V Velikina1, Leah C Henze Bancroft2, Alexey A Samsonov2, and James H Holmes2
1Department of Medical Physics, University of Wisconsin Madison, Madison, WI, United States, 2Department of Radiology, University of Wisconsin Madison, Madison, WI, United States
MOCCO reconstruction enables high spatial and temporal resolution DCE imaging for breast, which was proposed to be less sensitive to modeling error. However, the impact of noise level and lesion size on reconstruction performance is remained unknown. In this work we use a digital reference object phantom with the ability to adjust both spatial and temporal features for breast pharmacokinetic simulation. Further, we perform G-factor analysis of SNR performance, sharpness assessment, nRMSE and PK analysis for evaluating the temporal fidelity.
|
|
3460.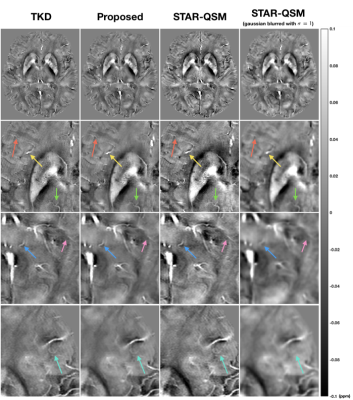 |
Sparse-Coding Regularized QSM Reconstruction for Suppressing Motion Artifacts
Jingjia Chen1 and Chunlei Liu1,2
1EECS, University of California, Berkeley, Berkeley, CA, United States, 2Helen Wills Neuroscience Institute, University of California, Berkeley, Berkeley, CA, United States
Subject motion downgrades QSM image quality and accuracy and can even nullify the image for diagnostic purposes in clinical settings. While QSM plays an emerging role in evaluating neurodegenerative diseases, motion artifact reduction is crucial for its adoption by researchers and clinicians. In this project, we develop a sparse-coding regularized QSM reconstruction algorithm to mitigate motion artifacts and noise. In vivo experiments suggest that the proposed method can alleviate motion artifacts to a certain extent while preserving sharp structures. This regularization technique can be applied jointly with other regularizations to achieve a desired susceptibility map.
|
3461.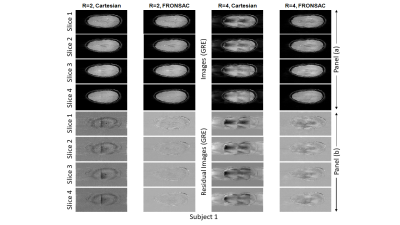 |
Quantitative comparison of a brain protocol with and without FRONSAC enhancement
E. H. Bhuiyan1, Y. Rodriguez1, R. Todd Constable1, and G. Galiana1
1Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States
FRONSAC has previously been demonstrated as a highly effective approach to using nonlinear gradients to reduce undersampling artifacts, but previous results were limited to proof-of-principle experiments in individual subjects. Here we report quantitative comparisons for a full protocol of standard brain imaging sequences (2D GRE, 2D TSE, 2D T2w-FLAIR, 3D GRE and 3D MP-RAGE) acquired in a cohort of healthy subjects, comparing standard Cartesian to FRONSAC-enhanced acquisitions. Preliminary results confirm that FRONSAC significantly improves undersampling artifacts, measured as RMSE relative to a fully sampled Cartesian reference, while retaining the contrast, SNR, and reliability of standard Cartesian sequences.
|
|
3462.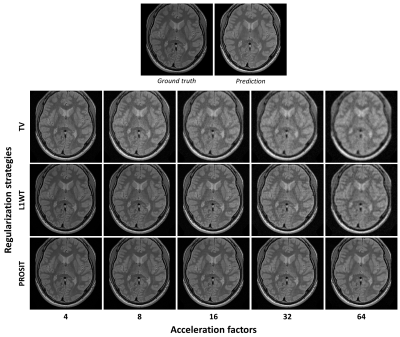 |
Contrast prediction-based regularization for iterative reconstructions (PROSIT)
Hendrik Mattern1, Alessandro Sciarra1,2, Max Dünnwald2,3, Soumick Chatterjee1,3,4, Ursula Müller1, Steffen Oeltze-Jafra2,5, and Oliver Speck1,5,6,7
1Biomedical Magnetic Resonance, Otto-von-Guericke University, Magdeburg, Germany, 2Medicine and Digitalization, Otto-von-Guericke University, Magdeburg, Germany, 3Faculty of Computer Science, Otto-von-Guericke University, Magdeburg, Germany, 4Data & Knowledge Engineering Group, Otto-von-Guericke-University, Magdeburg, Germany, 5Center for Behavioral Brain Sciences, Magdeburg, Germany, 6German Center for Neurodegenerative Disease, Magdeburg, Germany, 7Leibniz Institute for Neurobiology, Magdeburg, Germany
In this study, contrast prediction is used as an auxiliary tool to regularize underdetermined image reconstructions. This novel regularization strategy enables to share information across individual reconstructions and outperforms state of the art regularizations for high acceleration factors.
|
|
3463.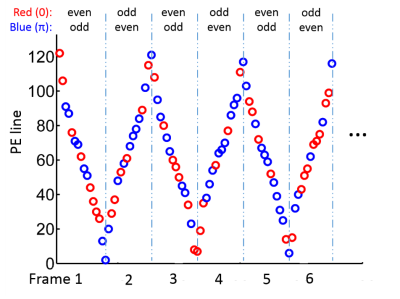 |
Highly Accelerated Free Breathing Whole Heart Real-time Cine using AuTO-calibrated Multiband Imaging and Compressed Sensing (ATOMICS)
Yu Ding1, Lele Zhao2, Zhongqi Zhang2, Qi Liu1, Jian Xu1, and Yuan Zheng1
1UIH America, Houston, TX, United States, 2United Imaging Healthcare, Shanghai, China
Conventional whole heart cine scan requires multiple breath-holds, and the prolonged scan time may limit its clinical use. We propose a highly accelerated whole heart real-time cine technique that combines AuTO-calibrated Multiband (MB) Imaging and Compressed Sensing (ATOMICS). With specially designed temporal phase modulation and in-plane undersampling pattern, single-band reference images can be extracted from the multiband data themselves, and cine images can be reconstructed using the CS framework. The proposed technique was demonstrated on a healthy volunteer with 16-fold acceleration (MB=2, undersampling factor=8), and free-breathing whole heart cine with diagnostic quality was acquired in about 12 seconds.
|
|
3464.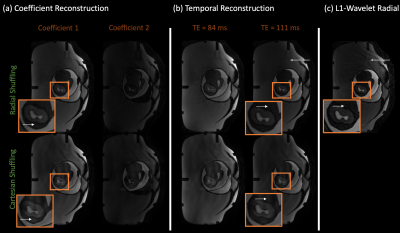 |
Improving cartesian single-shot 2D T2 shuffling and reducing radial streaking artifacts with golden angle radial T2 shuffling
Yamin Arefeen1, Fei Han2, Borjan Gagoski3,4, Jacob White1, and Elfar Adalsteinsson1,5,6
1Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 2MR R&D and Collaborations, Siemens Healthineers, Los Angeles, CA, United States, 3Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 4Harvard Medical School, Boston, MA, United States, 5Harvard-MIT Health Sciences and Technology, Cambridge, MA, United States, 6Institute for Medical Engineering and Science, Cambridge, MA, United States
T2 shuffling reduces blurring in 3D Fast Spin Echo (FSE) by incorporating a low rank representation of signal evolution in the forward model. However, extending T2 shuffling to 2D-FSE is challenging due to the limited spatiotemporal incoherence achievable with cartesian sampling. Radial trajectories offer increased incoherence in multiple dimensions in comparison to cartesian acquisitions. We combine golden angle radial sampling with the T2 shuffling forward model to improve image quality in comparison to 2D cartesian T2 shuffling and to reduce streaking artifacts in radial single-shot FSE images. Simulation and phantom experiments illustrate the advantages of radial T2 shuffling.
|
|
3465.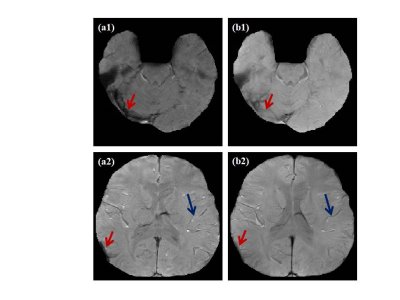 |
Enhancing GRE magnitude image using rank reduction of structured matrix
Sreekanth Madhusoodhanan1, Vazim Ibrahim1, Chandrasekharan Kesavadas2, and Joseph Suresh Paul1
1Indian Institute of Information Technology and Management-Kerala, Trivandrum, India, 2Sree Chitra Tirunal Institute for Medical Science and Technology, Trivandrum, India
The noise and phase dispersion in the multi-slice multi-echo Gradient Echo (GRE) acquisition causes non-exponential signal decay along the temporal dimension. Due to this, signal dropouts are observed in the air tissue interface. Construction of structured block Hankel matrix and rank minimization with a fidelity for data consistency reduces the signal loss and improves the visibility of venous structures in the GRE magnitude image.
|
|
3466.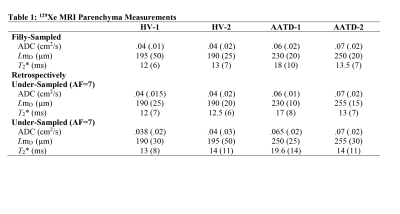 |
Can an Adapted SEM Accurately Deliver 129Xe MRI-based Lung Morphometry Estimates
Elise Noelle Woodward1, Matthew S Fox1,2, David G McCormack3, Grace Parraga3,4,5, and Alexei Ouriadov1,2
1Physics and Astronomy, Western University, London, ON, Canada, 2Lawson Health Research Centre, London, ON, Canada, 3Department of Medicine, Respiratory, Western University, London, ON, Canada, 4Robarts Research institute, London, ON, Canada, 5Medical Biophysics, Western University, London, ON, Canada
We hypothesize that the SEM equation can be adapted for fitting the spin-density dependence of the MR signal similar to fitting the time or b-value dependences: Signal=exp[-(nr)^β], where 0<β<1, n is an image number and r is the fractional spin-density. Such an approach permits consideration of the signal intensity variation as reflection of the underlying spin-density variation and hence, reconstruction of the under-sampled k-space using the adapted SEM equation. In this proof-of-concept evaluation, we have demonstrated the feasibility of this approach in a small group of patients. Lung SV/ADC/morphometry/T2* maps have been generated using reconstructed images and their corresponding weighing.
|
|
3467.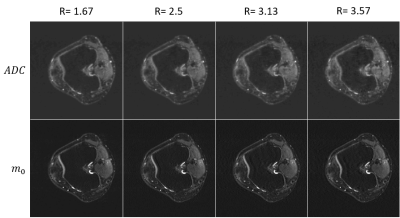 |
Model-Based Iterative Reconstruction for diffusion Data with Phase Correction and Penalty Term
Thomas Hüfken1, Jannik Arbogast1, Anna-Katinka Bracher2, Meinrad Beer2, Henning Neubauer2, and Volker Rasche 1
1Internal Medicine II, University Ulm Medical Center, Ulm, Germany, 2Department of Radiology, University Ulm Medical Center, Ulm, Germany
A diffusion model is used to generate a synthetic k-space which is compared with acquired k-space data. Minimization of the cost function with an additional regularization term in form of a total variation is performed by a nonlinear conjugated gradient solver. Phase alterations due to strong diffusion gradients are considered by introduction of a phase operator. Data of nine human knee joints were acquired with a multi-shot diffusion weighted EPI sequence. Undersampling factors between R=1.5 and R=3.57 yielded excellent results for magnitude and ADC data.
|
|
3468. |
Direct Estimation of pre-contrast T1 for DCE-MRI
Zhibo Zhu1, Yannick Bliesener1, R. Marc Lebel2,3, and Krishna Shrinivas Nayak1
1Electrical and Computer Engineering, University of Southern California, Los Angeles, CA, United States, 2Global MR Applications & Workflow, GE Healthcare, Calgary, AB, Canada, 3Radiology, University of Calgary, Calgary, AB, Canada
Quantitative DCE-MRI requires pre-contrast T1 mapping with matching resolution and coverage. Recent studies have shown that sparse sampling and constrained reconstruction can be applied to both DCE-MRI and pre-contrast T1 mapping with the variable flip angle (VFA) approach. Here, we demonstrate and evaluate direct estimation of T1 from sparsely sampled VFA raw data. In healthy subjects, we demonstrate superior T1 estimation compared with prior methods at subsampling factors >10.
|
|
3469.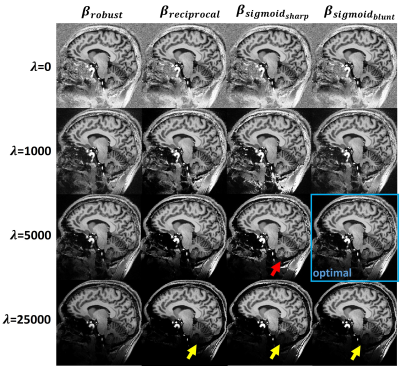 |
Near bias-free robust MP2RAGE reconstruction using a signal-dependent regularization function
Yi-Cheng Hsu1, He Wang2,3, and Ying-Hua Chu1
1MR Collaboration, Siemens Healthcare Ltd., Shanghai, China, 2Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China, 3Human Phenome Institute, Fudan University, Shanghai, China
MP2RAGE is a B1-insensitive T1-weighted imaging and T1 mapping method with strong background noise. Introducing a regularization parameter into the reconstruction can suppress the noise at the cost of lowering the signal intensity and T1 value. We propose a robust MP2RAGE using a signal-dependent regularization function. The reconstructed images are nearly bias-free with minimal artifact and reduced background noise.
|
|
3470. |
Online reconstruction of GRE Fat Navigators with Gadgetron on Siemens Terra 7T Scanner
Ayan Sengupta1,2, Iulius Dragonu3, and Christopher T. Rodgers2
1Department of Psychology, Royal Holloway, University of London, London, United Kingdom, 2Wolfson Brain Imaging Centre, University of Cambridge, Cambridge, United Kingdom, 3Siemens Healhineers, London, United Kingdom
High resolution Ultra High Field 7T imaging is highly prone towards involuntary motion artifacts. Fat Navigator based motion correction provides a robust solution but it is a retrospective correction method. In this study we present an online reconstruction method 3D GRE FatNavs with open-source reconstruction tool, Gadgetron. We improved the performance of the GRAPPA reconstruction pipeline in Gadgetron for fast online reconstruct FatNav. We also implemented a Python Gadget to perform fsl Flirt based co-registration through NiPype to produce motion parameters from the FatNav.
|
|
3471.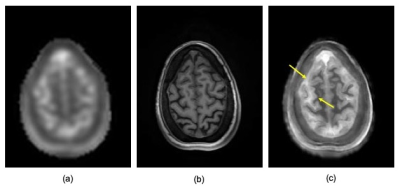 |
High Resolution Sodium MRI Through Manifold Approximation at 3T
Fernando Boada1, Hugh Wang1, Yongxian Qian1, Georg Schramm2, and Johan Nuyts2
1Radiology, New York University, New York, NY, United States, 2Katholieke Universiteit Leuven, Leuven, Belgium
We demonstrate the use of manifold approximation for implementing anatomically guided reconstructions for the removal of T2-related blurring from sodium images acquired at 3T.
|
|
3472.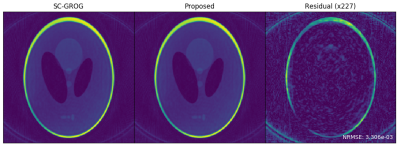 |
Prime Factorizations for Accelerated Radial SC-GROG
Nicholas McKibben1,2 and Edward V. DiBella1,2
1Biomedical Engineering, University of Utah, Salt Lake City, UT, United States, 2Radiology and Imaging Sciences, University of Utah, Salt Lake City, UT, United States
GROG is an attractive alternative to convolutional gridding and non-uniform DFT methods because of comparatively low cost and no density correction. However, for large multicoil datasets, many fractional matrix powers must be performed which scale with the cube of the number of channels. For SC-GROG and real-time SC-GROG, time and memory requirements can be significantly lowered for precomputation and updates of fractional powers by decomposing required powers into smaller, composable pieces. This is an NP-hard combinatorial change-making problem. We propose a simple solution based on prime factorization which leads to significant computational and memory savings with little performance degradation.
|
|
3473.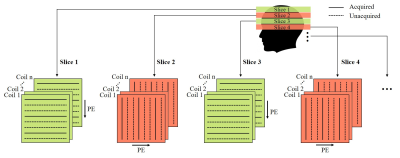 |
Calibration-free Highly Accelerated Multi-slice MRI via Alternating Phase Encoding Directions and Low-rank Tensor Completion Reconstruction
Yujiao Zhao1,2, Zheyuan Yi1,2,3, Yilong Liu1,2, Fei Chen3, Yanqiu Feng4, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong, China, 3Department of Electrical and Electronic Engineering, Southern University of Science and Technology, Shenzhen, China, 4School of Biomedical Engineering, Southern Medical University, Guangzhou, China
2D multi-slice MR data share strong correlations in coil sensitivities and image contents over adjacent slices. Here we’ve developed a new acquisition and reconstruction strategy for calibration-free multi-slice MRI. In brief, k-space data for each slice are uniformly undersampled along one phase encoding direction, while undersampled data for next adjacent slice are acquired with phase encoding along an orthogonal direction. Multiple images are then jointly reconstructed using a low-rank Hankel tensor completion approach. This method maximizes the incoherence of aliasing artifacts, and utilizes the coil sensitivity and image content correlations across adjacent slices, leading to high accelerations with uniform undersampling.
|
|
3474.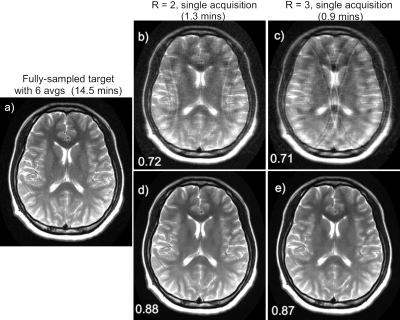 |
Sparse sampling reconstruction and noise removal of 0.55T brain MRI using a learned variational network
Patricia M. Johnson1, Zhang Le2, David Grodzki3, and Florian Knoll1
1Center for Biomedical Imaging, New York University, New york, NY, United States, 2Siemens Shenzhen Magnetic Resonance Ltd., Shenzhen, China, 3Siemens Healthcare GmbH, Erlangen, Germany
Most clinical MRI scanners operate at high magnetic field, however low-field MRI offers many advantages and promises to improve the value of MRI. The main drawback is low SNR; several signal averages are often required, which may result in prohibitively long scans. We can look to deep learning (DL) to facilitate accelerated low-field imaging through both denoising and sparse sampling. In this work, we use a variational network for both denoising and under-sampled reconstruction of brain images acquired on a 0.55T prototype system, demonstrating that low-field MRI paired with DL can produce high-quality images in very short scan times.
|
|
3475.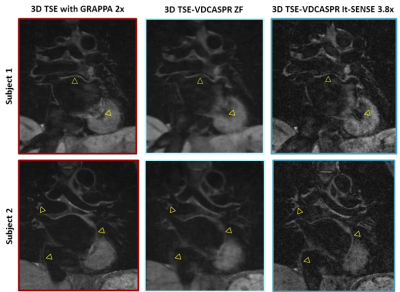 |
Accelerated Free-breathing 3D Whole-heart Black-blood TSE Imaging
Giovanna Nordio1, Radhouene Neji2, Karl Kunze2, Rene Botnar1, and Claudia Prieto1
1Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom
In this study we investigate the feasibility of a 3D turbo spin-echo imaging technique with variable flip angle combined with an undersampled Cartesian variable density trajectory (3D TSE-VDCASPR) for efficient whole-heart black-blood imaging. Data from five healthy subjects are acquired with the proposed 3D TSE-VDCASPR and the conventional 3D TSE with Cartesian GRAPPA. The proposed imaging 3D TSE-VDCASPR imaging technique allows for comparable black-blood imaging, with the advantage of reduce nominal scan time (4.6±1 vs 8.1±1.6 minutes ± seconds). Future work will investigate more advanced motion compensation and image reconstruction techniques in order to achieve predictable and fast scan time.
|
|
3476.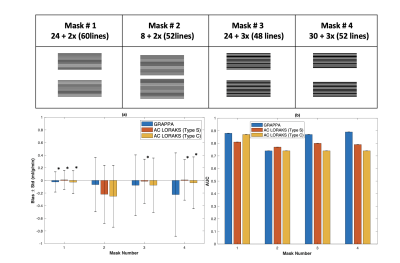 |
Optimal data sampling and image reconstruction for Cartesian bSSFP ASL-CMR
Erum Mushtaq1, Ahsan Javed1, and Krishna S. Nayak1
1University of Southern California (USC), Los Angeles, CA, United States
Arterial spin labeled cardiovascular magnetic resonance (ASL-CMR) is a non-contrast myocardial perfusion imaging technique, which can detect angiographically significant coronary artery disease. Sensitivity of ASL-CMR can be improved by shortening the imaging window with parallel imaging which reduces physiological noise from cardiac motion. In this work, we explore the optimal Cartesian sampling pattern and parallel imaging reconstruction strategy for bSSFP ASL-CMR. We consider different under-sampling masks, acceleration factors, and reconstruction techniques to minimize the imaging window. The optimal setting is selected based on the bias in MP measurements.
|
3477.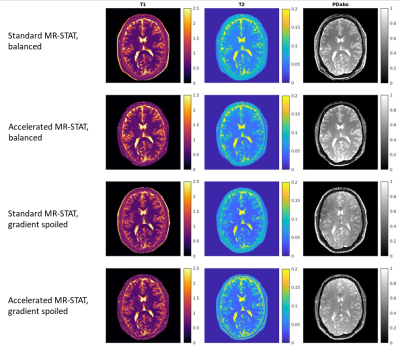 |
Accelerated MR-STAT Algorithm: Achieving 10-minute High-Resolution Reconstructions on a Desktop PC
Hongyan Liu1,2, Oscar van der Heide1,2, Cornelis A.T. van den Berg1,2, and Alessandro Sbrizzi1,2
1Computational Imaging Group for MR Diagnostics and Therapy, Center for Image Sciences, University Medical Center Utrecht, Utrecht, Netherlands, 2Department of Radiology, Division of Imaging and Oncology, University Medical Center Utrecht, Utrecht, Netherlands
MR-STAT is a framework for simultaneous mapping of quantitative MR parameters from a single short scan. Since MR-STAT involves the solution of a large scale nonlinear optimization problem, the reconstruction time has always been one main concern. In the current work, we develop an accelerated MR-STAT algorithm, which achieves two order of magnitude acceleration in reconstruction times. High-resolution 2D dataset can be reconstructed within 10 minutes on a desktop PC thereby drastically facilitating the application of MR-STAT in the clinical work-flow.
|
|
3478.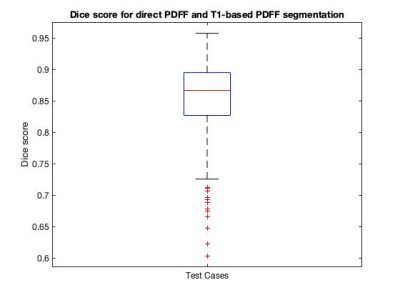 |
Automatic segmentation for liver fat quantification
Elisabeth Sarah Pickles1,2, Alexandre Bagur1,2, Ged Ridgway2, Benjamin Irving2, Daniel Bulte1, and Michael Brady2,3
1Institute of Biomedical Engineering, Oxford University, Oxford, United Kingdom, 2Perspectum Diagnostics, Oxford, United Kingdom, 3Department of Oncology, Oxford University, Oxford, United Kingdom
By segmenting the liver on an MRI Proton Density Fat Fraction (PDFF) map a median PDFF value is obtained, indicating the amount of fat in the liver. Automatic segmentation is desirable, as manual segmentation is time consuming. We investigated a direct PDFF automatic segmentation method using a U-Net model and compared it to a T1-based PDFF segmentation. We show that the median values obtained are comparable, and the Dice scores are relatively good, although not as high as desired. Visually the direct PDFF segmentation is not always optimal. We suggest that improvement of the model is desirable.
|
|
3479.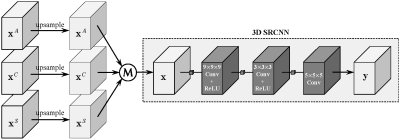 |
Isotropic MRI Reconstruction with 3D Convolutional Neural Network
Xiaole Zhao1, Tian He1, Ying Liao1, Yun Qin1, Tao Zhang1,2,3, and Mark Zou1,2,3
1School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, China, 2High Field Magnetic Resonance Brain Imaging Laboratory of Sichuan, Chengdu, China, 3Key Laboratory for NeuroInformation of Ministry of Education, Chengdu, China
Typical magnetic resonance imaging (MRI) usually shows distinct anisotropic spatial resolution in imaging plane and slice-select direction. Image super-resolution (SR) techniques are widely used as an alternative method to isotropic MRI reconstruction. In this work, we propose to reconstruct isotropic magnetic resonance (MR) volumes via 3D convolutional neural network in an end-to-end manner. 3D SRCNN is utilized to preliminarily validate the idea and it produces quantitative and qualitative results significantly superior to traditional methods, such as Cube-Avg and NLM methods.
|
|
3480.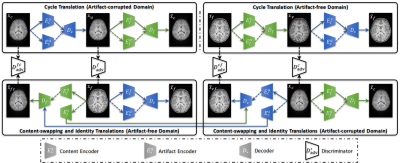 |
Retrospective Artifact Correction of Pediatric MRI via Disentangled Cycle-Consistency Adversarial Networks
Siyuan Liu1, Kim-Han Thung1, Weili Lin1, Pew-Thian Yap1, and the UNC/UMN Baby Connectome Project Consortium2
1Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 2University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Retrospective artifact removal using supervised learning requires explicit generation of artifact-corrupted images and is impractical since generating the wide variety of potential artifacts can be challenging. Using unsupervised learning, we show how artifacts can be disentangled with remarkable efficacy from artifact-corrupted images to recover the artifact-free counterparts, without requiring explicit artifact generation.
|
|
3481.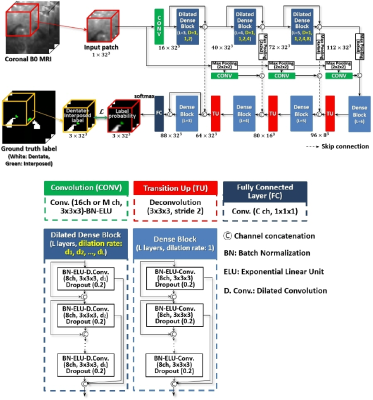 |
Deep Cerebellar Nuclei Segmentation via Semi-Supervised Deep Context-Aware Learning from 7T Diffusion Image
Jinyoung Kim1, Rémi Patriat1, Jordan Kaplan1, and Noam Harel1
1Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States
In this study, we proposed the first deep learning and 7T MR imaging based dentate and interposed nuclei segmentation framework. We introduce dilated dense blocks to effectively encode contextual information on different receptive fields in an encoder-decoder network. Training of the proposed network is optimized with a multi-class hybrid segmentation loss, handling a class imbalance problem. Moreover, a self-training strategy facilitates the training of the proposed network by exploiting auxiliary labels. The proposed framework significantly outperforms an atlas-based deep cerebellar nuclei segmentation tool and state-of-the-art deep neural networks in terms of accuracy and consistency.
|
|
3482.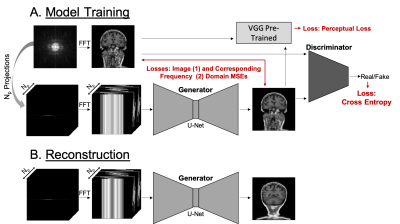 |
PROJECTION GAN: Highly accelerated projection reconstruction using generative adversarial neural network
Tetiana Dadakova1, Jian Wu1, Hyun-Kyung Chung1, Brian Anhalt1, Dmitry Tkach1, Alexander Graff1, Natalie Marie Schenker-Ahmed1, David Karow1, and Christine Leon Swisher1
1Human Longevity, Inc., San Diego, CA, United States
Many clinical MRI applications in chest and abdomen require low sensitivity to motion. In addition, high acquisition speed is necessary for imaging in non-cooperative patients or those unable to perform breath holds. These applications would benefit from the highly accelerated radial acquisition. Deep learning has been shown to provide good results for image reconstruction from highly under-sampled k-space data. Here we introduce a Projection GAN - a generative adversarial neural network, which is trained to reconstruct highly accelerated MR images from uniformly rotated projections. Our results show that even with aggressive under-sampling the reconstruction has great overall performance.
|
|
3483.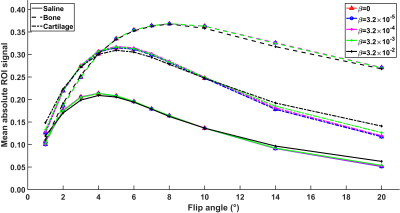 |
Quantitative 3-D SWIFT with compressed sensing
Antti Paajanen1, Olli Nykänen1, Matti Hanhela1, Nina Hänninen1,2, Swetha Pala1, Ville Kolehmainen1, and Mikko J Nissi1
1Department of Applied Physics, University of Eastern Finland, Kuopio, Finland, 2Research Unit of Medical Imaging, University of Oulu, Oulu, Finland Poster Permission Withheld
Despite the extra information offered by the quantitative magnetic resonance imaging (qMRI), these methods are not widely used due to their long acquisition times. Our approach for fast qMRI was built on minimized data acquisition with ultra-short echo time imaging, and compressed sensing image reconstruction. To validate the approach, a set of variable flip angle images of equine osteochondral specimens were acquired with the Multi-Band-SWIFT sequence. The 4-D image stack was reconstructed with CS-framework utilizing spatial and contrast regularizations. Results with 81% reduced data showed comparable image quality while maintaining correct contrast modulation for quantitative parameter (T1 relaxation time) estimation.
|
|
3484.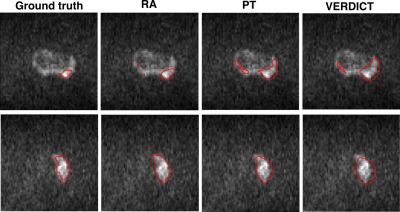 |
Domain adaptation for prostate lesion segmentation on VERDICT-MRI
Eleni Chiou1,2, Francesco Giganti3,4, Shonit Punwani5, Iasonas Kokkinos2, and Eleftheria Panagiotaki1,2
1Centre of Medical Imaging Computing, University College London, London, United Kingdom, 2Department of Computer Science, University College London, London, United Kingdom, 3Department of Radiology, UCLH NHS Foundation Trust, University College London, London, United Kingdom, 4Division of Surgery & Interventional Science, University College London, London, United Kingdom, 5Centre for Medical Imaging, Division of Medicine, University College London, London, United Kingdom
The successful adoption of convolutional neural networks (CNNs) for improved diagnosis can be hindered for pathologies and clinical settings where the amount of labelled training data is limited. In such cases, domain adaptation provides a viable alternative. In this work we propose domain adaptation to enhance the performance of prostate lesion segmentation on VERDICT-MRI utilising diffusion weighted (DW)-MRI data from multi-parametric (mp)-MRI acquisitions. Experimental results show that domain adaptation significantly improves the segmentation performance on VERDICT-MRI.
|
|
3485.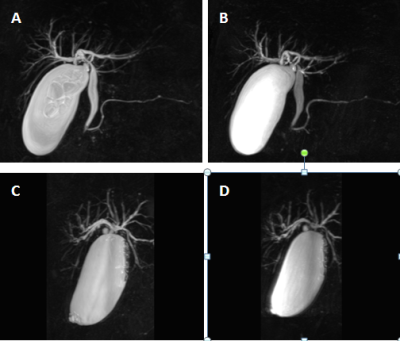 |
Rapid 3D Navigator-Triggered MRCP With SPACE Sequence at 3T: Only One-third Acquisition Time of Conventional 3D Navigator-Triggered MRCP.
Zhiyong Chen1, bin sun1, yunjing xue1, zhongshuai zhang2, and guijin li3
1Radiology, Union Hospital, Fujian Medical University, Fuzhou, China, 2Diagnostic imaging, Siemens Healthcare, Shanghai, China., shanghai, China, 3MR application, Siemens Healthineers Ltd,Guangzhou,China, guangzhou, China The SPACE sequence based rapid MRCP protocol proposed in this study, which reduced acquisition time without deteriorating the image quality, yielded significantly higher overall image quality and better visualization of the pancreaticobiliary tree compared with the conventional MRCP. On the basis of our findings, we suggest that the rapid 3D SPACE technique could improve the clinical throughput of MRCP and show a trend toward wider clinical availability for MRCP studies. |
|
| 3486. | WITHDRAWN | |
3487.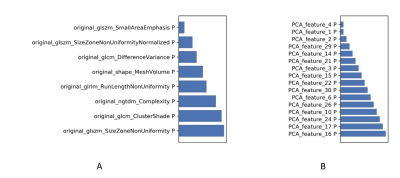 |
Comparing the performance of optimized support vector machine model in predicting efficacy of chemo-radiotherapy for advanced rectal cancer with and without a paired-difference up-sampling strategy under small sample size
Jie Kuang1, QingLei Shi2, Gaofeng Shi1, Xu Yan3, and LI Yang1
1The Fourth Hospital of Hebei Medical University, Shijiazhuang, China, 2Siemens Healthcare, MR Scientific Marketing, Beijing, China, 3Siemens Healthcare, MR Scientific Marketing, Shanghai, China
In this study, we evaluated the effect of paired-difference analysis (PDA) up-sampling strategy on the performance of optimized support vector machine model (SVM) in predicting efficacy of chemo-radiotherapy for advanced rectal cancer. A higher accuracy and robustness was gained for the model adopted the PDA method in predicting the efficacy of treatment, which means that the PDA method can be used as an up-sampling strategy in improving the performance of some machine learning models.
|
|
3488.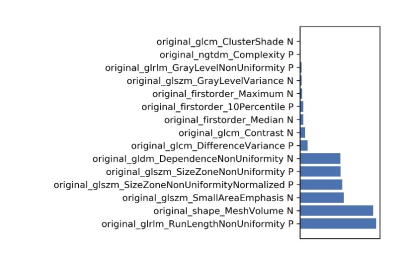 |
Predicting the efficacy of neoadjuvant chemo-radiotherapy for advanced rectal cancer using random forest model and a paired-difference up-sampling strategy under small sample size
Jie Kuang1, QingLei Shi2, Gaofeng Shi1, Xu Yan3, and LI Yang1
1The Fourth Hospital of Hebei Medical University, Shijiazhuang, China, 2Siemens Healthcare, MR Scientific Marketing, Beijing, China, 3Siemens Healthcare, MR Scientific Marketing, Shanghai, China
In this study, we adopted a paired-difference strategy, which can improve training efficiency of random forest (RF) model with a small sample size. Through optimizing in normalization, dimensional reduction, and features selection steps, a higher accuracy was achieved in predicting the efficacy of chemo-radiotherapy for advanced rectal cancer.
|
|
3489.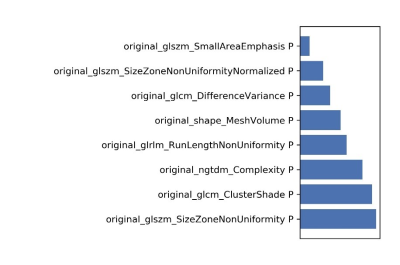 |
Predicting the efficacy of chemo-radiotherapy for advanced rectal cancer using support vector machine model and a paired-difference up-sampling strategy under small sample size
Jie Kuang1, Xu Yan2, Gaofeng Shi1, QingLei Shi3, and LI Yang1
1The Fourth Hospital of Hebei Medical University, Shijiazhuang, China, 2Siemens Healthcare, MR Scientific Marketing, Shanghai, China, 3Siemens Healthcare, MR Scientific Marketing, Beijing, China
In this study, we proposed a paired-difference analysis (PDA) method to up-sample the training data, which can improve training efficiency of support vector machine (SVM) model with a small sample size. Through optimizing in normalization, dimensional reduction, and features selection steps, a high accuracy was achieved in predicting the efficacy of chemo-radiotherapy for advanced rectal cancer, which means that valuable information may be provided and showed in this clinical situation.
|
|
3490.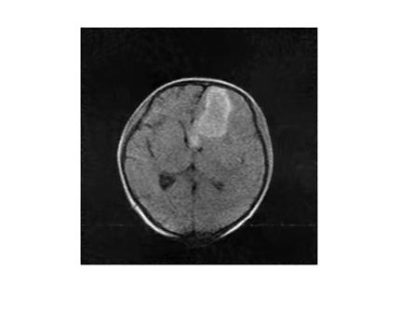 |
Low-rank based compressed sesning in low-field MRI for stroke
Fangge Chen1, Zheng Xu1, Yucheng He1, and Liang Tan2
1State Key Laboratory of Power Transmission Equipment and System Security and New Technology, Chongqing University, Chongqing, China, 2Department of Neurosurgery, Southwest Hospital, Chongqing, China
While being of great worth in convenience and timely scanning for severe disease, low-field and accessible MRI suffer from long scanning time. To speed up the scanning process in low-field MRI, a low-rank based compressed sensing method with a non-convex reconstruction model is proposed and solved by weighted SVT and gradient descent method iteratively. The in vivo data acquired from a Hemorrhage patient at a 0.05T MRI scanner is used for simulation. The reconstructed image (20% sampling rate) reveals the same hemorrhage shape as CT image shows, demonstrating the ability of compressed sensing applied in low-field MRI cliniclly.
|
|
3491.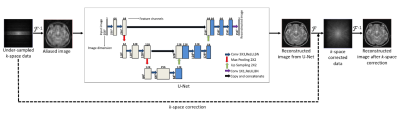 |
Deep Learning based Accelerated MR Image Reconstruction via Transfer Learning
Madiha Arshad1, Mahmood Qureshi1, and Hammad Omer1
1Department of Electrical and Computer Engineering, COMSATS University, Islamabad, Pakistan
In MRI, many deep learning-based solutions often degrade when deployed in different clinical scenarios due to lack of large training datasets. Moreover, the knowledge about the reconstruction problem is constrained to the data seen during training. This paper presents a transfer learning approach to deal with the problems of data scarcity and differences in the source and target domain while reconstructing the MR images using deep learning. Experimental results show successful domain transfer between the source and target datasets in terms of change in magnetic field strength, anatomy and Acceleration Factor (AF).
|
|
3492.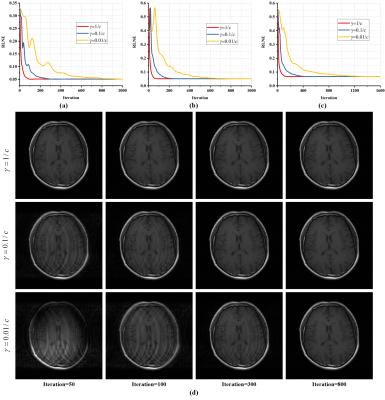 |
A Convergence Proof of Projected Fast Iterative Soft-thresholding Algorithm for Parallel Magnetic Resonance Imaging
Xinlin Zhang1, Hengfa Lu1, Di Guo2, Lijun Bao1, Feng Huang3, and Xiaobo Qu1
1Department of Electronic Science, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Fujian Provincial University Key Laboratory of Internet of Things Application Technology, Xiamen University of Technology, Xiamen, China, 3Neusoft Medical System, Shanghai, China
For compressed sensing magnetic resonance imaging, algorithms plays a significant role in sparse reconstruction. The Projected Fast Iterative Soft-thresholding Algorithm (pFISTA), a simple and efficient algorithm solving sparse reconstruction models, has been successfully extended to parallel imaging. However, its convergence criterion still poses as an open question, yielding no guideline for parameter setting that allows faithful results. In this work, we prove the sufficient conditions for the convergence of parallel imaging version of pFISTA. Results on in vivo data demonstrate the validity of the convergence criterion.
|
3493.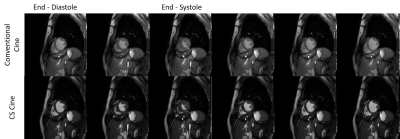 |
Use of Compressed Sensing to Reduce Scan Time and Breath-holding for cine bSSFP MRI in Children and Young Adults
Nivedita Naresh1, Ladonna Malone1, Takashi Fujiwara1, Emma Hulseberg-Dwyer1, Janet McGee1, Quin Lu2, Mark Twite3, Michael Dimaria4, Brian Fonseca4, Lorna P Browne1, and Alex J Barker1,5
1Radiology, Children's Hospital Colorado, Aurora, CO, United States, 2Philips Healthcare North America, San Francisco, CA, United States, 3Anesthesiology, Children's Hospital Colorado, Aurora, CO, United States, 4Pediatrics, Children's Hospital Colorado, Aurora, CO, United States, 5Bioengineering, University of Colorado Anschutz Medical Campus, Aurora, CO, United States
In this study, we have a compared a vendor-optimized compressed sensing (CS) accelerated cine MRI technique with the conventional cine MRI technique. There were no significant differences in image quality and cardiac volumes between the two techniques, except left ventricular end-diastolic volume which was significantly lower with the CS technique. The accelerated technique on average reduced scan time by 50%.
|
|
3494.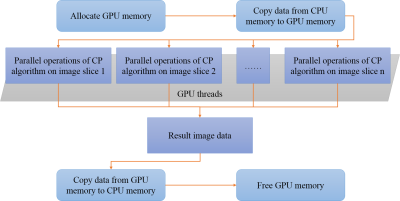 |
Accelerating Positive Contrast MRI Reconstruction with GPU-based Parallel Chambolle-Pock Algorithm
Fang Cai1, Caiyun Shi1, Jing Cheng1, Guoxi Xie2, Hanwei Chen3, Xin Liu1, Hairong Zheng1, Dong Liang1, and Haifeng Wang1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Department of Biomedical Engineering, Guangzhou Medical University, Guangzhou, China, 3Department of Radiology, Guangzhou Panyu Central Hospital, Guangzhou, China
Positive contrast Magnetic Resonance Imaging (MRI) based on the susceptibility mapping requires very fast reconstruction for clinical applications. Modern graphics processing units (GPUs) are very efficient at manipulating computer graphics and image processing. And Chambolle-Pock(CP) algorithm is also an efficient algorithm to solve the minimization problem. To further reduce the reconstruction time of positive contrast MRI, a GPU-based parallel CP algorithm was proposed. The experimental results showed that the proposed GPU-based parallel CP method could achieve similar image results, and provide a faster reconstruction of up to 15 times than the conventional CPU-based CP method.
|
|
3495.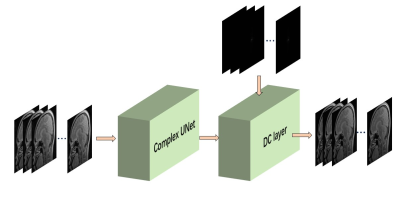 |
Weakly Supervised Deep Prior Learning for Multi-coil MRI Reconstruction
Haoyun Liang1, Taohui Xiao1, Chuyu Rong1, Yu Gong1, Cheng Li1, and Shanshan Wang1
1Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, Chinese Academy of Sciences, Shenzhen, P.R.China, Shenzhen, China
MRI reconstruction based on supervised learning methods, have achieved remarkable success. However, because of the difficulty and high cost of the MR images collection process, it is not always easy to obtain a big dataset with strong supervised information. Therefore, weakly supervised learning will be a possible solution. In this work, we proposed a weakly deep prior learning algorithm to train a complex UNet for multi-coil MR images reconstruction without very precise labels. The result shows that our proposed algorithm can provide a competitive performance compared to the classical methods with enoucraging quantitative indicators of SSIM and PSNR.
|
|
3496.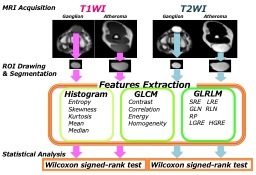 |
Imaging biomarker to extract features from soft tissue tumors
Kazuya Kishi1, Daisuke Yoshimaru2, Yasuwo Ide3, and Keito Saitou4
1Department of radiology, Chiba medical center, Chiba, Japan, 2RIKEN Center for Brain Science, Wako, Japan, 3Department of radiology, Chiba Central Medical Center, Chiba, Japan, 4Department of Medical Technology, Tokyo Women's Medical University Yachiyo Medical Center, Yachiyo, Japan
In order to evaluate the pathological alteration in soft tissue tumors, we focused on the color, distribution, and morphology of the images, based on the tumor components. We could classify soft tissue tumors by texture analysis using MRI. T2-weighted images were more useful for classifying these tumors than T1-weighted images. Texture analysis was extremely useful for diagnosis of soft tissue tumors.
|
|
3497.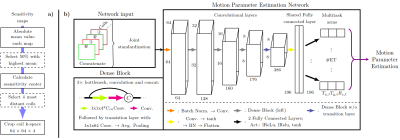 |
MoPED: Motion Parameter Estimation DenseNet for accelerating retrospective motion correction
Julian Hossbach1,2,3, Daniel Nicolas Splitthoff2, Stephen Farman Cauley4, and Andreas Maier1
1Pattern Recognition Lab, Friedrich-Alexander-University Erlangen-Nuremberg, Erlangen, Germany, 2Siemens Healthcare GmbH, Erlangen, Germany, 3Erlangen Graduate School in Advanced Optical Technologies, Erlangen, Germany, 4Martinos Center for Biomedical Imaging, Charlestown, MA, United States
We exploit the data redundancy and the locality of motion in k-space for an estimation of the motion parameters using a Deep Learning approach. The exploratory Motion Parameter Estimation DenseNet (MoPED) extracts the in-plane motion parameters between echo trains of a TSE sequence. As input, the network receives the center patch of the k-space from multiple coils; the network’s output can serve multiple purposes. While an image rejection/reacquisition can be triggered by the motion guess, we show that motion aware reconstruction can be accelerated using MoPED.
|
|
3498.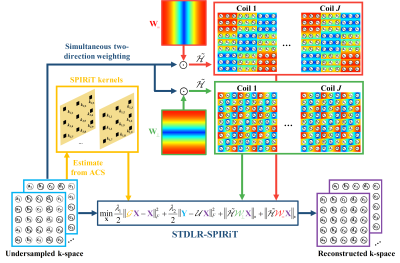 |
Image Reconstruction with Low-rankness and Self-consistency of k-space Data in Parallel MRI
Xinlin Zhang1, Di Guo2, Yiman Huang1, Ying Chen1, Liansheng Wang3, Feng Huang4, and Xiaobo Qu1
1Department of Electronic Science, Fujian Provincial Key Laboratory of Plasma and Magnetic Resonance, Xiamen University, Xiamen, China, 2School of Computer and Information Engineering, Fujian Provincial University Key Laboratory of Internet of Things Application Technology, Xiamen University of Technology, Xiamen, China, 3Department of Computer Science, School of Information Science and Engineering, Xiamen University, Xiamen, China, 4Neusoft Medical System, Shanghai, China
Recent low-rank reconstruction methods offer encouraging image reconstruction results enabling promising acceleration of parallel magnetic resonance imaging, however, they were not originally designed to exploit the routinely acquired calibration data for performance improvement in parallel magnetic resonance imaging. In this work, we proposed an image reconstruction approach to simultaneously explore the low-rankness of the k-space data and mine the data correlation among multiple receiver coils with the use of the calibration data. The proposed method outperforms the state-of-the-art methods in terms of suppressing artifacts and achieving lowest error, and exhibits robust reconstructions even with limited auto-calibration signals.
|
|
3499.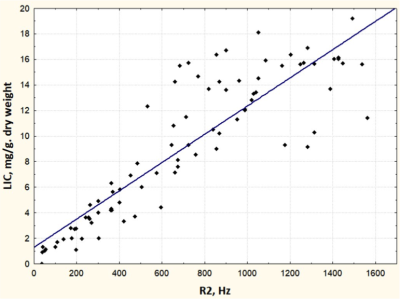 |
Free breath T2*-mapping for planning and monitoring the chelation therapy in children with secondary iron overload
Evelina Nazarova1, Galina Tereshchenko1, and Dmitry Kupriyanov2
1Radiology, Dmitry Rogachev National Medical Research Center Of Pediatric Hematology, Oncology and Immunology, Moscow, Russian Federation, 2Philips Healthcare, Imaging Systems, Moscow, Russian Federation
Iron overload represents critical health problem in children with regular transfusional therapy. Breath hold T2* mapping is widely used for the iron quantification, but it is not always feasible in pre-school children without the sedation. We purpose the free breath T2* mapping for the quick monitoring of treatment response to the chelation therapy in pediatric patients.The purposed method demonstrate that free breath T2* mapping is more convenient for the iron quantification in children according to lack of limited by breath hold technique and provides much higher quality images.
|
|
3500.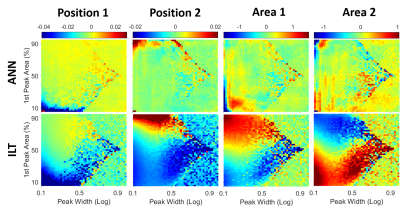 |
Continuous T2 Distribution Analysis using Artificial Neural Networks
Tristhal Parasram1, Rebecca Daoud1, and Dan Xiao1
1University of Windsor, Windsor, ON, Canada
Quantitative analysis of T2 relaxation times could reveal molecular scale information, and has significance in the study of brain, spinal cord, articular cartilage, and cancer discrimination. However, it is non-trivial to recover the relaxation times from MR signals, especially as a continuous distribution, since it is an intrinsically ill-posed problem. In this work, ANNs have been trained to generate the T2 distribution spectra. The performance was evaluated across a large parameter range. In addition to superior computation speed, higher accuracy was achieved compared to the traditional method.
|
|
3501.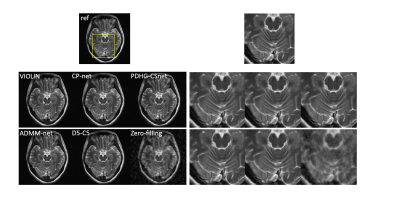 |
VIOLIN: improVIng mOdel-based deep learning reconstruction by reLaxing varIable combiNations
Jing Cheng1, Yiling Liu1, Qiegen Liu2, Ziwen Ke1, Haifeng Wang1, Yanjie Zhu1, Leslie Ying3, Xin Liu1, Hairong Zheng1, and Dong Liang1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Nanchang University, Nanchang, China, 3University at Buffalo, The State University of New York, Buffalo, Buffalo, NY, United States
Most of the unrolling-based deep learning fast MR imaging methods learn the parameters and regularization functions with the network architecture structured by the corresponding optimization algorithm. In this work, we introduce an effective strategy, VIOLIN and use the primal dual hybrid gradient (PDHG) algorithm as an example to demonstrate improved performance of the unrolled networks via breaking the variable combinations in the algorithm. Experiments on in vivo MR data demonstrate that the proposed strategy achieves superior reconstructions from highly undersampled k-space data.
|
|
3502.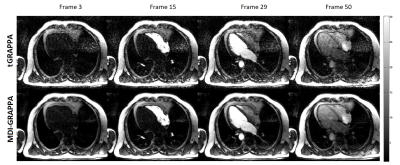 |
Improving Dynamic Paralleling Imaging Reconstruction using Multi-Dimensional Integration (MDI)
Jingyuan Lyu1, Yongquan Ye1, Sen Jia2, Zheng Shi3, Zhongqi Zhang4, Lele Zhao4, Jian Xu1, and Rui Yang3
1UIH America, Inc., Houston, TX, United States, 2Paul C. Lauterbur Research Centre for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 3Henan Chest Hospital, Zhengzhou, China, 4United Imaging Healthcare, Shanghai, China
We have developed and demonstrated a novel strategy for accurate estimation of coil sensitivity profiles for time-resolved data. Instead of direct temporal average to reduce noise in ACS, the proposed multi-dimensional integrated (MDI) strategy solves the least square problem for coil sensitivity profiles estimation in a new fashion. By extracting the overall calibration equations in all temporal dimensions, MDI is able to calculate coil sensitivity profiles more accurately than without this strategy. It can also be integrated with other parallel imaging methods such as SENSE and ESPIRiT.
|
|
3503.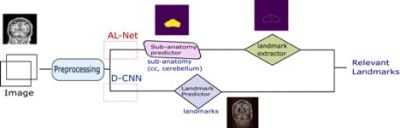 |
Deep Convolutional Neural Network model for sub-Anatomy specific Landmark detection on Brain MRI
Sumit Sharma1 and Srinivasa Rao Kundeti1
1Philips Healthcare, Bangalore, India
A Deep CNN (D-CNN) model for Brain sub-anatomy landmark detection for auto MRI scan planning. We compare D-CNN with traditional approaches like segmentation followed by image processing (AL-Net). D-CNN shows better landmark detection with Average RMSE <= 6mm (N=100) compared to AL-Net.
|
|
3504.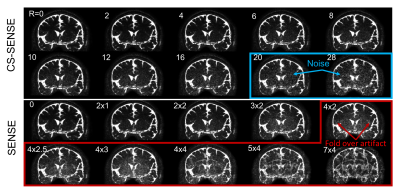 |
Evaluation of point-spread-function and signal-to-noise ratio in highly-accelerated Compressed Sensing-SENSE (CS-SENSE) and SENSE MRI
Di Cao1,2,3, Adrian G.Paez1,2, Xinyuan Miao1,2, Dapeng Liu1,2, Chunming Gu1,2,3, and Jun Hua1,2
1F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 2The Russell H. Morgan Department of Radiology and Radiological Sciences, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States
We seek to experimentally evaluate the point-spread-function (PSF) and signal-to-noise ratio (SNR) in sequences accelerated using conventional SENSE and the recently developed Compressed Sensing-SENSE (CS-SENSE) with acceleration-factors (R) ranging from 0 to 28. Both CS-SENSE and SENSE had little effect on the PSF in the tested 3D turbo-spin-echo (TSE) sequences. CS-SENSE showed preserved SNR-per-unit-time even when R=28 (compared to R=0), while SENSE reduced SNR-per-unit-time significantly when R≥4. Fold-over artifacts were seen on SENSE images with R≥8, but not on CS-SENSE images for R=0-28. Overall, CS-SENSE seems to show clear advantages compared to SENSE, especially with high acceleration-factors.
|
|
3505.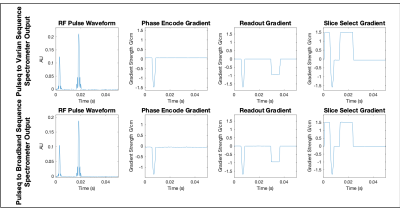 |
Integration of Open Source Pulse Sequence Programming Toolbox, Pulseq, for use with System Agnostic Broadband Spectrometer
Courtney Bauer1 and Steven M. Wright1
1Electrical and Computer Engineering, Texas A&M University, College Station, TX, United States
Introduced is a new translator for the open source pulse sequence programming toolbox, Pulseq. Unique from previous translator units developed, this translator takes the Pulseq output file and generates a time-bin based, delimited pulse sequence table. This format is specific to the target system, a homebuilt broadband spectrometer. Our lab uses multiple scanners, and using Pulseq enables seamless transition between a conventional research scanner, a Varian Unity Inova or a prototype homebuilt spectrometer. Ultimately, this translator is not only a useful tool, but it introduces the possibility of facilitating a direct comparison of homebuilt systems with their commercial counterparts.
|
|
3506.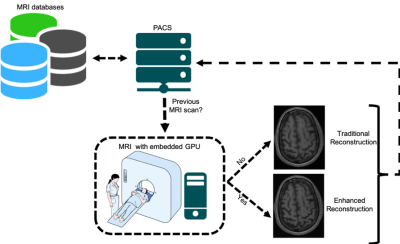 |
Enhanced Deep-learning-based Magnetic Resonance Image Reconstruction using Subjects’ Previous Scans
Roberto Souza1, Youssef Beauferris1, Wallace Loos1, Mariana Bento1, Robert Marc Lebel2, and Richard Frayne1
1University of Calgary, Calgary, AB, Canada, 2GE, Calgary, AB, Canada
Magnetic resonance (MR) compressed sensing reconstruction explores image sparsity to make MR acquisition faster while still reconstructing high quality images. Modern picture archiving and communication systems allow efficient access to previous scans acquired of the same subject. In this work, we propose to use previous scans to enhance the reconstruction of follow-up scans using a deep learning model. Our model is composed of a reconstruction network that outputs an initial MR reconstruction, which is used as input to an enhancement network along with a co-registered previous scan. Our enhancement network improved quantitative metrics on average by 15%.
|
|
3507.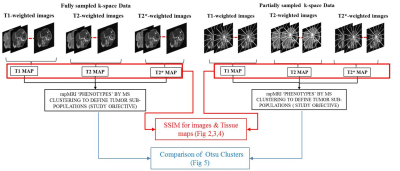 |
Joint Reconstruction of Multiparameter MRI Images for Imaging Intratumoral Subpopulations
Shraddha Pandey1,2, Arthur David Snider1, Wilfrido Moreno1, and Natarajan Raghunand2
1Electrical Engineering, University of South Florida, Tampa, FL, United States, 2Cancer Physiology, Moffitt Cancer Research Center, Tampa, FL, United States
Multispectral analysis of multiparametric MRI (mpMRI) images is being explored as an approach to objectively quantify intratumoral heterogeneity. However, acquisition of multiple co-registered parameters maps such as T1, T2, T2* and ADC for this task can be time-consuming. We propose a joint reconstruction method that exploits shared structural information between co-registered MRI parameter maps that provides high similarity between multispectral clusters defined on parameter maps from undersampled data and the ground truth fully-sampled maps of M0, T1, T2 and T2*.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Video
Watch the Video View the Poster
View the Poster Back to Top
Back to Top