Digital Poster Session
Acquisition, Reconstruction & Analysis: Software Tools and Image Analysis
Acquisition, Reconstruction & Analysis
3644 -3659 Software Tools and Image Analysis - Software Tools
3660 -3672 Software Tools and Image Analysis - Image Analysis 1
3673 -3688 Software Tools and Image Analysis - Image Analysis 2
Session Topic: Software Tools and Image Analysis
Session Sub-Topic: Software Tools
Digital Poster
Acquisition, Reconstruction & Analysis
3644.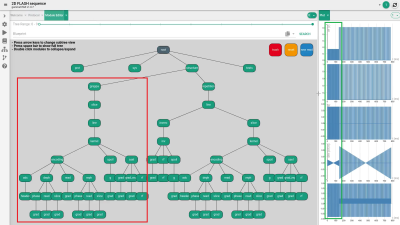 |
Graphical sequence visualization and development in the dynamic platform-independent framework gammaSTAR (γ*): A first prototype
Simon Konstandin1, Cristoffer Cordes1, and Matthias Günther1,2
Video Permission Withheld
1MR Physics, Fraunhofer MEVIS, Bremen, Germany, 2MR-Imaging and Spectroscopy, Faculty 01 (Physics/Electrical Engineering), University of Bremen, Bremen, Germany
MR sequence development is usually carried out by means of manufacturer-specific frameworks that do not allow an easy sequence transfer to scanners from other manufacturers. Sequence development in the recently presented framework is performed by scripting that can be time consuming and confusing for sophisticated MR sequences. Here, a module editor prototype is presented that provides a graphical sequence representation and should allow for a fast and relatively easy-to-understand MR sequence development in the future. Functionality could already be demonstrated by exchanging the rf pulse and implementing a parallel acquisition technique into a gradient echo sequence in a few steps.
|
|
3645.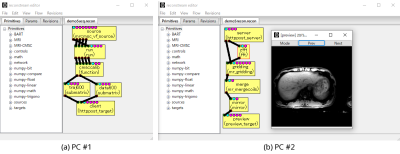 |
Reconstream: A Software Framework for Developing Image Reconstruction Algorithms in Scientific Research Activities
Hidenori Takeshima1
1Advanced Technology Research Department, Research and Development Center, Canon Medical Systems Corporation, Kanagawa, Japan
Existing software tools for assisting scientific research mainly focus on increasing reproducibility of primitives and/or integrated algorithms (exchangeable reproducibility). While exchangeable reproducibility is clearly important, reproducing the outputs using past configurations again is also important in daily research activities (local reproducibility).
The author proposes a new software framework named reconstream. To improve local reproducibility, reconstream implemented a revision manager for reproducing past configurations easily. To improve exchangeable reproducibility, the author reviewed advantages of individual software tools and implemented them in the reconstream framework. Experimental results demonstrate several examples using various features of reconstream. |
|
3646.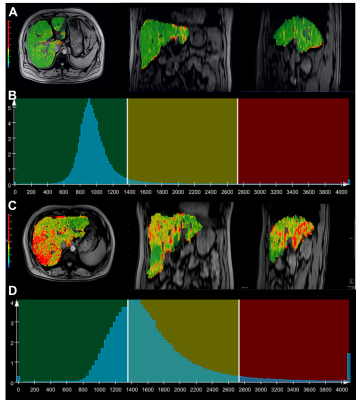 |
Utility of Whole-Liver Histogram and Texture Analysis on T1 Maps in the Risk Stratification of Advanced Fibrosis in NAFLD
Xinxin Xu1, Caixia Fu2, Robert Grimm3, Huimin Lin1, Ruokun Li1, and Fuhua Yan1
1Radiology, Ruijin Hospital affiliated to Shanghai Jiaotong University School of Medicine, Shanghai, China, 2Siemens healthcare, Shanghai, China, 3Siemens healthcare, Erlangen, Germany
We aim to assess the utility of whole-liver texture analysis on T1 maps for the risk stratification of advanced fibrosis in patients with suspected NAFLD. Our experiments show that whole-liver histogram and texture parameters of T1 maps can distinguish NAFLD patients with an intermediate-to-high risk of advanced fibrosis. A combination of histogram and texture parameters as a multivariate model showed a better diagnostic performance than any sole parameter and noninvasive fibrosis test in risk stratification of advanced fibrosis.
|
|
3647.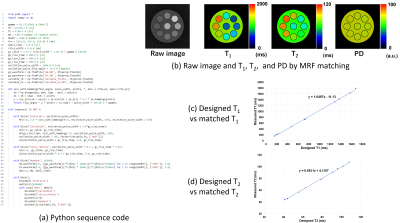 |
Python pulse sequence development kit for a fast MRI simulator
Ryoichi Kose1 and Katsumi Kose1
1MRIsimulations,Inc., Tokyo, Japan
A software tool that can efficiently describe MRI pulse sequences in Python has been developed for a fast MRI simulator. The essential part of this tool consists of application program interface (API) developed in C++, and any pulse sequence can be written by calling the API from a Python program. Using this tool, several pulse sequences including magnetic resonance fingerprinting were developed, and their usefulness was evaluated by performing MRI simulations for numerical phantoms. As a result, the tool developed in this study was shown to be very efficient in developing MRI pulse sequences.
|
|
3648.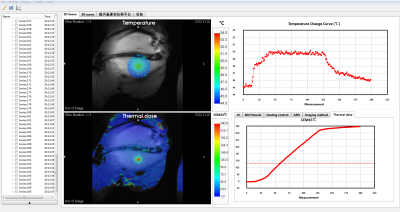 |
An MR guided focused ultrasound software with Gadgetron reconstruction
Yangzi Qiao1, Chao Zou1, Jianhong Wen1, Sen Jia1, Xin Liu1, and Hairong Zheng1
1Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China
An integrated focused ultrasound guidance software named MARFit was developed base on the framework of Gadgetron. The software realized automatically focus localization and real-time temperature change monitoring during HIFU therapy. These features has been evaluated in animal experiments. The software makes the post processing procedure easily translated between different vendors, and could be a powerful tool for MR guided focused ultrasound therapy.
|
|
3649.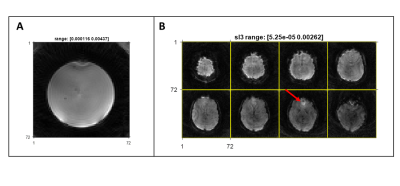 |
Cross-vendor implementation of a Stack-of-spirals PRESTO BOLD fMRI sequence using TOPPE and Pulseq
Marina Manso Jimeno1,2, Sairam Geethanath1,2, Jon-Fredrik Nielsen3, and Douglas C. Noll3
1Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center (CMRRC), New York, NY, United States, 3University of Michigan, Ann Arbor, MI, United States
TOPPE and Pulseq are two examples of open-source, vendor-agnostic frameworks for rapid prototyping and implementation of MR sequences. Both have demonstrated their ability to easily execute a variety of highly specific and customized sequences on different vendors. Cross-vendor translation of sequences generated by these two frameworks can enable multi-site and multi-vendor evaluation of a single sequence implementation. This study demonstrates this capability by porting a spiral-based fMRI sequence implemented in TOPPE on one vendor platform to Pulseq to run on another vendor. Results show comparable in vivo SNR and image contrast for Pulseq and TOPPE implementations.
|
|
3650.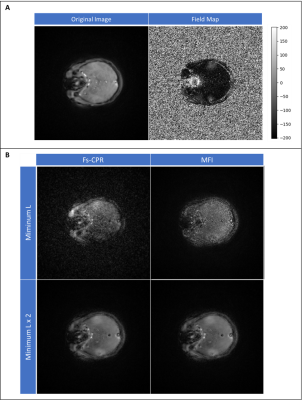 |
Open-source Python package for spiral off-resonance correction
Marina Manso Jimeno1,2, Sairam Geethanath1,2, and John Thomas Vaughan Jr.1,2
1Columbia University, New York, NY, United States, 2Columbia Magnetic Resonance Research Center (CMRRC), New York, NY, United States
Off-resonance blurring is one of the challenges with long read-out trajectories such as spirals. Existent techniques deblur the images by demodulating k-space data with the conjugate term of the phase accumulated from off-resonant spins. Continuous and frequency-segmented CPR and Multi-Frequency Interpolation are some examples. This work is aimed at sharing an open-source package for off-resonance correction, demonstrating on spiral datasets. The methods have been tested on simulations, in vitro and in vivo and show promising preliminary results. We believe that the availability of this package will benefit the spiral imaging community.
|
|
3651.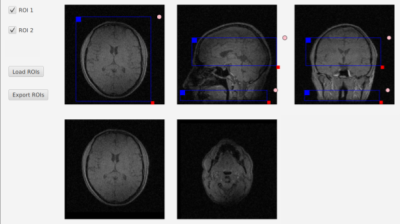 |
An open-source graphical tool for interactive slice planning: Application to pseudo-continuous Arterial Spin Labeling
Jon-Fredrik Nielsen1, Luis Hernandez-Garcia1, and Douglas C. Noll1
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Vendor-agnostic MR pulse sequence programming tools such as Pulseq or TOPPE allow rapid prototyping of complex MR sequences, but are not compatible with existing graphical slice planning interfaces on all supported vendor platforms. We introduce a simple open-source slice planning tool that allows any number of 3D rectangular regions-of-interest (ROIs) to be defined interactively and exported to a portable data file format (HDF5). We use this tool to prescribe both a 2D labeling plane in the neck and a 3D imaging volume in the brain for a vendor-agnostic implementation of 3D stack-of-spirals fast spin-echo (FSE) pseudo-continuous Arterial Spin Labeling.
|
|
3652.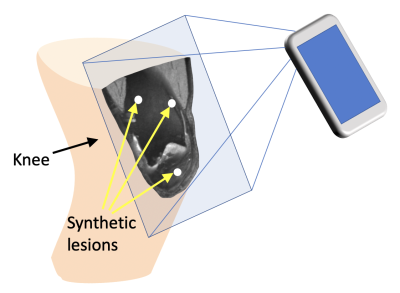 |
Intuitive MRI Data Visualization and Reslicing Using Augmented Reality
Bragi Sveinsson1,2, Neha Koonjoo1,2, and Matthew Rosen1,2
1Massachusetts General Hospital, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States
We present a smartphone app to view 3D medical image data using Augmented Reality (AR). The app superimposes the imaged anatomy on the patient and allows for completely free and intuitive choice of viewing plane by rotating the phone and moving it towards and away from the patient. The app is shown to be enable free choice of viewing plane in a much easier and faster manner than standard medical image viewing software on a computer workstation. We envision this app to enable lower-cost, more mobile viewing of MRI data, potentially in combination with portable, low-field MR hardware.
|
|
3653.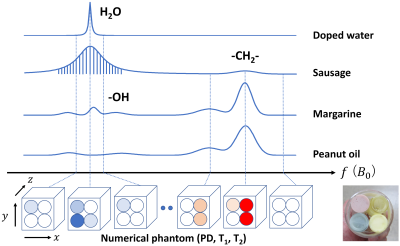 |
Development of a method for Bloch image simulation of living tissues
Katsumi Kose1, Ryoichi Kose1, Yasuhiko Terada2, Daiki Tamada3, and Utaroh Motosugi3
1MRIsimulations, Tokyo, Japan, 2University of Tsukuba, Tsukuba, Japan, 3University of Yamanashi, Chuo, Japan
A method for Bloch image simulation of living tissues was developed and the simulated results were compared with experiments performed for phantoms including T2* distribution and various chemical components. The Bloch image simulation for living tissues is based on a four-dimensional numerical phantom consisting of spatially defined 3D datasets of proton density, T1, and T2 with spatially homogeneous B0 (frequency) offset. The multiple gradient-echo images reconstructed from the MR signal obtained from the Bloch image simulation of the numerical phantom reproduced experimental results for samples that simulated living tissues.
|
|
3654.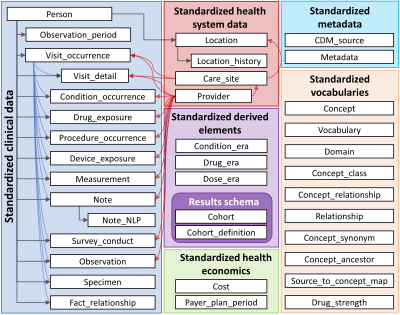 |
Development of a management system for radiology–common data model (R-CDM) and its application in the liver disease: extension of OMOP-CDM
Min-Gi Pak1, Seong-Min Han2, ChungSub Lee3, SeungJin Kim1, Tae-Hoon Kim3, Chang-Won Jeong3, and Kwon-Ha Yoon3,4
1Medical Science, Wonkwang University, Iksan, Republic of Korea, 2Computer Software Engineering, Wonkwang University, Iksan, Republic of Korea, 3Medical Convergence Research Center, Wonkwang University, Iksan, Republic of Korea, 4Radiology, Wonkwang University, Iksan, Republic of Korea Poster Permission Withheld
The Observational Medical Outcomes Partnership-Common Data Model (OMOP-CDM) used in distributed research networks has low coverage of clinical data and does not reflect the latest trends of precision medicine. Radiology data have great merits to visual and identify the lesions in specific diseases. However, radiology data should be shared to obtain the sufficient scale and diversity required to provide strong evidence for improving patient care. Our study was to develop a web-based management system for radiology-CDM (R-CDM), as an extension of the OMOP-CDM, and to evaluate the feasibility of R-CDM dataset for application of radiological image data in clinical practice.
|
|
3655. |
Assessing generalized multi-pool exchange tissue model MRI simulations for the modelling of an ultra-low field scanner
Ignacio Xavier Partarrieu1, Elliot Fox1,2, Frederic Brochu1, Matt Cashmore1, Neha Koonjoo3, Sheng Shen3, David Sinden1, Matthew Rosen3, and Matt G. Hall1,4
1National Physical Laboratory, Teddington, United Kingdom, 2Durham University, Durham, United Kingdom, 3Harvard, Boston, MA, United States, 4University College London, London, United Kingdom
The large expense and running costs of traditional 1.5T scanners have meant that as demand for scans has grown supply has not been able to keep up. Novel, ultra-low field systems aim to address this problem by producing images with some resolution losses as a trade-off for reduced costs. However, access to such devices is currently restricted, limiting the possible research output. In this work we develop and test a simulation model of 6.5 mT b-SSFP acquisitions, which we compare to actual scanner acquisitions. We find that such simulations produce qualitatively and quantitatively similar images to real scans.
|
|
3656.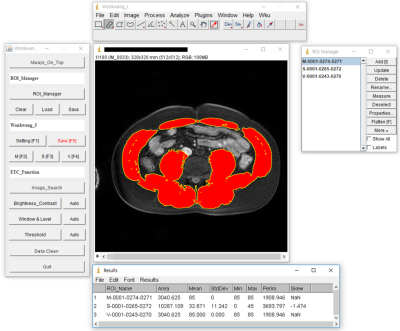 |
Development of a quantification software for body composition imaging and its assessment in the patient with sarcopenia
SeungJin Kim1, Min-Gi Pak1, Tae-Hoon Kim2, Chang-Won Jeong2, and Kwon-Ha Yoon2,3
1Medical Science, Wonkwang University, Iksan, Republic of Korea, 2Medical Convergence Research Center, Wonkwang University, Iksan, Republic of Korea, 3Radiology, Wonkwang University, Iksan, Republic of Korea Poster Permission Withheld
In 2016, sarcopenia has been classified by the international classification of diseases(ICD-10-CM), with the code(M62.84). The role of imaging techniques has rapidly increased in the field of sarcopenia. In the past decade, the importance of muscle and fat mass has been emphasized on imaging evaluation of body composition including of muscle and body fat such as visceral fat or subcutaneous fat. However, there are diverse quantification methods for assessing muscle and fat mass by imaging and thus, these methods must be standardized. This study developed a customized quantification software based on ImageJ-platform and evaluated in the patient with sarcopenia.
|
|
3657.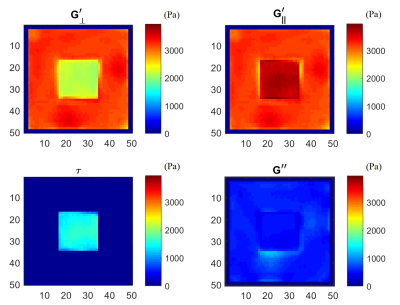 |
Development and validation of a novel finite element inversion method and analysis pipeline for anisotropic MR Elastography
Behzad Babaei1, Daniel Fovargue2, David Nordsletten2,3, and Lynne Bilston1,4
1Neuroscience Research Australia, Sydney, Australia, 2King's College London, London, United Kingdom, 3Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 4Prince of Wales Clinical School, University of New South Wales, Randwick, Australia
MR elastography is well established for measuring the mechanical properties of soft tissues in a broad range of clinical populations. However, unlike liver tissue, which is generally considered to be isotropic, muscle is significantly anisotropic, with stiffer mechanical behaviour along the muscle fibre directions. Identifying anisotropic properties is challenging due to the need to track fibre directions and additional numerical difficulties in estimating properties. Here, we describe a novel finite element inversion method for estimating anisotropic mechanical properties of ex vivo tissue data, and validate this method using three sets of simulated data.
|
|
3658.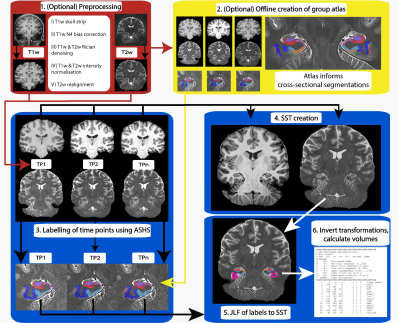 |
Automatic Longitudinal Assessment of Hippocampal Subfields using Multi-Contrast MRI
Thomas B Shaw1, Steffen Bollmann1, Ashley York1, Maryam Ziaei1, and Markus Barth1
1The University of Queensland, Brisbane, Australia
Characterising longitudinal changes in the subfields of the hippocampus using high-resolution MRI is important for understanding neurological disorders and diseases including Alzheimer’s. Hippocampus subfields are differentially involved in these disorders and can be used as biomarkers for disease. Here, we introduce an automatic longitudinal pipeline for measuring the subfields of the hippocampus (LASHiS). Unlike other longitudinal pipelines, LASHiS harnesses multiple MR contrasts and is therefore more sensitive to changes in these subfields. We compared LASHiS with four other established methods including Freesurfer and found that our method yields higher test-retest reliability and robust Bayesian longitudinal linear mixed effects modelling results.
|
|
3659.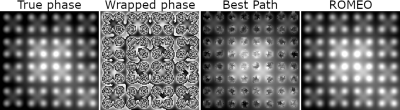 |
Rapid Opensource Minimum Spanning TreE AlgOrithm for Phase Unwrapping (ROMEO)
Simon Daniel Robinson1,2,3, Korbinian Eckstein1, Siegfried Trattnig1, Karin Shmueli4, and Barbara Dymerska4
1Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Department of Neurology, Medical University of Graz, Graz, Austria, 3Centre for Advanced Imaging, University of Queensland, Queensland, Australia, 4University College London, London, United Kingdom
We present a phase unwrapping algorithm (ROMEO), which improves on previous path-based unwrapping methods by using weights (the values of which determine the order in which voxels are unwrapped) that guide the unwrapping on paths through the object which avoid noise and a computationally efficient minimum spanning tree algorithm. ROMEO was tested against the region-growing method PRELUDE and the voxel-based method Best Path in unwrapping simulated data (a complex topography) and in-vivo GRE and EPI data acquired at 7T. ROMEO was found to be faster and more reliable than PRELUDE and Best Path.
|
Session Topic: Software Tools and Image Analysis
Session Sub-Topic: Image Analysis 1
Digital Poster
Acquisition, Reconstruction & Analysis
3660.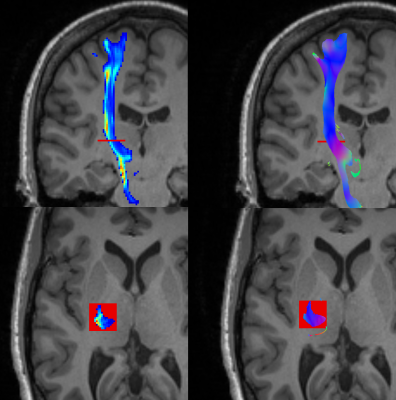 |
Measures of lesion load on white matter fiber bundles for damage assessment
Guillem Garcia1, David Moreno-Dominguez1, Marc Ramos 1, and Matt Rowe1
1Medical Imaging, QMENTA Inc., Boston, MA, United States
Measures of lesion load are useful to study how damage in the white matter present in different pathologies relate to changes in cognitive function. So far, most research focus on global or local volumetric metrics, an approach that exhibits limitations in cases where small lesions in specific places cause major damages. In this work, we propose the combined use of a set of metrics that measure different aspects of the lesions over the major white matter bundles. We expose how these metrics provide complementary information, and discuss how its usefulness could be assessed in future work.
|
|
3661.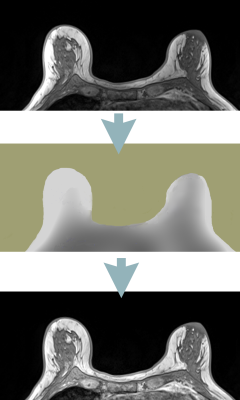 |
Retrospective MRI non-uniformity correction: quantitative assessment of two methods
Artem Mikheev1, Louisa Bokacheva1, Heesoo Yang1, Jeremy Sobel1, Carlos Fernandez-Granda2, Hersh Chandarana1, and Henry Rusinek1
1Radiology, NYU School of Medicine, New York City, NY, United States, 2Courant Institute of Mathematical Science, New York University, New York City, NY, United States
We have implemented a method (BiCal) for correction of image nonuniformity. Using objective criteria we have compared BiCal to widely used N4 algorithm in several challenging clinical MRI applications. There was a significant advantage of BiCal over N4 for 7T brain MRI and for accelerated radial GRASP of the abdomen. The performance of BiCal and N4 were comparable in breast imaging.
|
|
3662.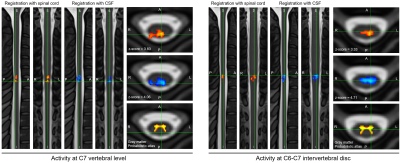 |
Improving spatial normalization of functional MRI data of the spinal cord using cerebrospinal fluid segmentation
Benjamin De Leener1,2, Linda Soltrand Dahlberg2, Ali Khatibi2,3, Nawal Kinany2,4, and Julien Doyon2
1Department of computer engineering and software engineering, Polytechnique Montreal, Montreal, QC, Canada, 2Montreal Neurological Institute, McGill University, Montreal, QC, Canada, 3Center of Precision Rehabilitation for Spinal Pain (CPR Spine), University of Birmingham, Birmingham, United Kingdom, 4Center for Neuroprosthetics, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland
Analyzing functional MRI of the spinal cord is challenging due to large susceptibility artifacts and inter-subject variability in terms of shape and curvature. More particularly, the poor contrast between the spinal cord and cerebrospinal fluid (CSF) leads to co-registration errors when analyzing the spinal cord functional activity at the group level. This study proposes a new registration framework that leverages the contrast between the CSF and its surrounding structure. Results show an increase of 67% in the number of active voxels at the group level, with an increase of 5% and 23% for the mean and max z-score, respectively.
|
|
3663.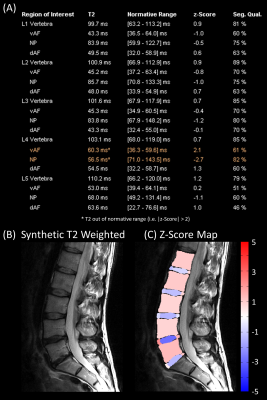 |
Fully Automated Quantitative Assessment of the Lumbar Intervertebral Disk
Tom Hilbert1,2,3, Marcus Raudner4, Markus Schreiner4,5, Anna Szelenyi4, Vladimir Juras4, Siegfried Trattnig4, and Tobias Kober1,2,3
1Advanced Clinical Imaging Technology, Siemens Healthcare, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4High Field MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 5Department of Orthopaedics and Trauma Surgery, Medical University of Vienna, Vienna, Austria
Quantitative imaging can detect subtle changes invisible to the naked eye and enables establishing healthy normative ranges. We report a comprehensive pipeline for fully automated assessment of the lumbar intervertebral disk based on a single fast T2-mapping acquisition of 2:27 min. In this proof-of-concept study, norm values are derived from 19 healthy subjects. Feasibility is shown in 19 patients: three regions are automatically segmented in each intervertebral disk and compared to corresponding norm values. Results are then compiled in a report where abnormal regions are flagged.
|
|
3664.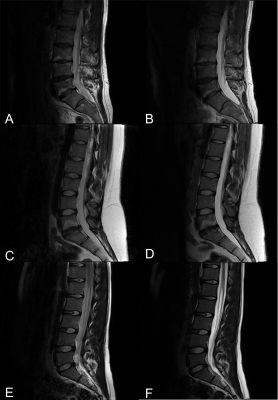 |
Synthetic T2-weighted contrasts of the lumbar spine derived from fast quantitative mapping – the icing on the cake
Marcus Raudner1, Markus Schreiner1,2, Tom Hilbert3,4,5, Tobias Kober3,5,6, Anna Szelenyi7, Vladimir Juras1, and Siegfried Trattnig1
1High Field MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Department of Orthopedics and Trauma Srugery, Medical University of Vienna, Vienna, Austria, 3Advanced Clinical Imaging Technology, Siemens Healthcare, Lausanne, Switzerland, 4Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 5LTS5, Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 6Department of Radiology, University Hospital and University of Lausanne, Lausanne, Switzerland, 7Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria
GRAPPATINI has already been shown to allow short acquisition times with diagnostic synthetic T2-weighted (T2w) images in the knee. This study investigates how different effective echo times influence the resulting contrast-to-noise ratio of GRAPPATINI compared to a conventional T2w turbo spin-echo (TSE) sequence in the spine. Overall, the synthetic T2w images maintained diagnostic quality while being reconstructed from highly-undersampled k-space data in 2:27 min acquisition time vs. 2:01 min for the T2w TSE with the advantage of GRAPPATINI to also offer multiple synthetic contrasts alongside robust T2 mapping.
|
|
3665.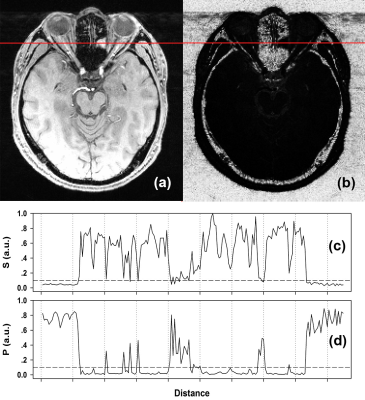 |
Complex semi-quantitative noise analysis for single set of MR images
Yongquan Ye1, Jingyuan Lv1, Yichen Hu1, Zhongqi Zhang2, Jian Xu1, and Weiguo Zhang1
1UIH America, Inc., Houston, TX, United States, 2United Imaging Healthcare, Shanghai, China
We propose a noise analysis method based on complex MRI signals. By actively inducing concomitant fluctuation into a single complex MR image, an abrupt change in a predefined signal function (e.g. signal ratio) is observed when signal intensity becomes stronger than background noise. The method is very sensitive to the existence of weak signal such as tissue-background boundaries or very low signal tissues, and can be used to semi-quantitatively distinghuish regions of signal and noise.
|
|
3666.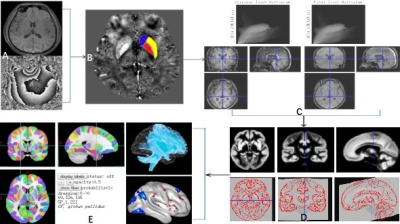 |
Evaluation of a standardized quantitative method of brain Iron based on quantitative susceptibility mapping and Brainnetome Atlas
Dongxue Li1, Bin Dai1, Zhenliang Xiong1, Xianchun Zeng1, Lisha Nie2, Pu-Yeh Wu2, and Rongpin Wang1
1Department of Radiology, Guizhou Provincial People's Hospital, Guiyang, China, 2GE Healthcare, MR Research, Beijing, China
Delineating the region of interest manually to obtain the magnetic susceptibility values from quantitative susceptibility mapping (QSM) for diagnosis of neurological diseases is a common method. However, this approach is time-consuming and may be inaccurate due to human error. Here we introduced a standardized brain region susceptibility quantification procedure based on QSM and Brainnetome Atlas. Our results show that this method has high accuracy in measuring brain iron content. Based on this method, we found that the content of brain iron tends to increase with age. Additionally, magnetic susceptibility values of partial brain regions have sex differences should be noted.
|
|
3667.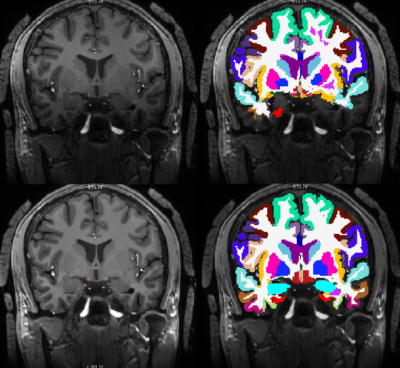 |
Optimizing a preprocessing pipeline for structural 7T MR analyses in FreeSurfer
Giske Opheim1,2, Oula Puonti3,4, Jan Ole Pedersen5, Vincent O. Boer3, Ane Kloster1,2, Martin Prener1,2, Helle Juhl Simonsen6, Olaf B. Paulson1,2, Lars H. Pinborg1,2, and Melanie Ganz1,7
1Neurobiology Research Unit, Rigshospitalet, Copenhagen University Hospital, Copenhagen, Denmark, 2Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark, 3Danish Research Centre for Magnetic Resonance, Funktions- og Billeddiagnostisk Enhed, Copenhagen University Hospital Hvidovre, Copenhagen, Denmark, 4Dept. of Health Technology, Technical University of Denmark, Kongens Lyngby, Denmark, 5Philips Healthcare, Copenhagen, Denmark, 6Functional Imaging Unit, Department of Clinical Physiology, Nuclear Medicine and PET, Rigshospitalet Glostrup, Copenhagen, Denmark, 7Dept. of Computer Science, University of Copenhagen, Copenhagen, Denmark
Automated cortical segmentations benefit from higher SNR and spatial resolutions on 7T MR images, but are also challenged by B1 inhomogeneities, causing faulty surface inflations primarily in the temporal lobes. We investigated how FreeSurfer outputs were affected by applying eight different preprocessing schemes prior to reconstructions of submillimeter 7T MPRAGE images. The highest segmentation robustness across subjects was obtained by setting bias-field correction FWHM to 60mm and adding light regularization, and additionally performing intensity normalization.
|
|
3668.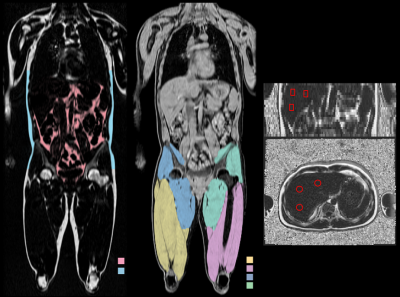 |
MRI-based Body Composition Analysis – Reproducibility and Repeatability
Magnus Borga1,2, André Ahlgren2, Thobias Romu2, Per Widholm2,3, Olof Dahlqvist Leinhard2,3, and Janne West2,3
1Department of Biomedical Engineering and Center for Medical Image Science and Visualization (CMIV), Linköping University, Linkoping, Sweden, 2AMRA Medical, Linköping, Sweden, 3Department of Medicine and Health and Center for Medical Image Science and Visualization (CMIV), Linköping University, Linköping, Sweden
This study investigated between-scanner reproducibility and repeatability of a method for MRI-based body composition analysis. Eighteen healthy volunteers were scanned twice on five different MR scanners from three vendors including both 1.5 and 3T, using a 2-point Dixon neck-to-knee imaging protocol. Visceral adipose tissue, abdominal subcutaneous adipose tissue, thigh muscle volume, muscle fat infiltration and liver fat were quantified using AMRA® Researcher. The reproducibility coefficients for the volume measurements ranged from 7 cl (anterior thigh muscles) to 4.2 dl (abdominal subcutaneous adipose tissue). The reproducibility coefficient for muscle fat infiltration was 1.5 percentage points and for liver fat 2.4 pp.
|
|
3669.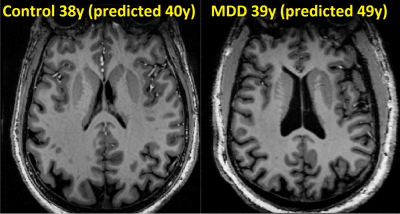 |
Quantification of brain age using high-resolution 7T MR imaging and implications in major depressive disorder
Gaurav Verma1, Yael Jacob1, Laurel Morris2, Priti Balchandani1, and James Murrough2
1Radiology, Icahn School of Medicine at Mount Sinai, NEW YORK, NY, United States, 2Psychiatry, Icahn School of Medicine at Mount Sinai, NEW YORK, NY, United States
A regression model estimating brain-age from about 250 T1-weighted imaging features was developed using data from 29 healthy controls (mean age 39.8). The model estimated brain age with average absolute error of 6.0 years. The model was applied 35 patients (mean age 38.7) diagnosed with major depressive disorder (MDD), yielding similar performance (7.6 years mean absolute error), but showed trend of over-estimation of average brain-age by 2.4 years. This technique demonstrates the feasibility of brain-age estimation using imaging features, and may help assess the differential effects of pathology like MDD in the aging process.
|
|
3670.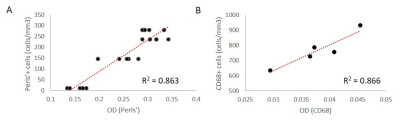 |
Color deconvolution and optical density of Multiple Sclerosis lesions
Jerry Kang1,2, Kelly Gillen1, and Yi Wang1
1Weill Cornell Medicine, New York, NY, United States, 2Bronx High School of Science, Bronx, NY, United States
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system characterized by focal inflammatory demyelination. In chronic active lesions, microglia and macrophages may contain high amounts of iron and express markers of pro-inflammatory polarization, driving tissue damage and disease progression. Therefore, studying the mechanisms behind iron accumulation and microglial inflammation are of great clinical importance, but require rigorous histological characterization of autopsied brain tissue from MS patients. We developed a rapid, automated method to quantify histopathological markers in human tissue and then validated our findings with manual counting and quantitative susceptibility mapping (QSM).
|
|
3671.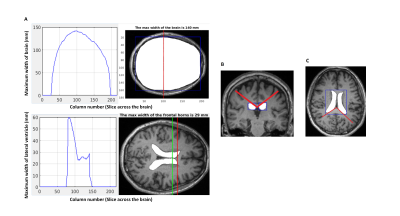 |
Differentiating Predominant Gait Disorder Parkinsonism Using Automated Geometric Indices
Ling Yun Yeow1, Bhanu Prakash KN1, AJY Lee2, EK Tan3,4, and LL Chan2,4
1Signal Image Processing Group, Singapore BioImaging Consortium, A*STAR, Singapore, Singapore, 2Diagnostic Radiology, Singapore General Hospital, Singapore, Singapore, 3National Neuroscience Institute – SGH Campus, Singapore, Singapore, 4Duke-NUS Medical School, Singapore, Singapore, Singapore Poster Permission Withheld
Gait apraxia has attributed to ventriculomegaly and periventricular leukoariaosis. Geometric quantification of ventriculomegaly using Evans’ Index (EI) and Callosal Angle (CA) have been proposed as biomarkers of normal pressure hydrocephalus (NPH). The novel Splenial Angle (SA) may aid in differentiating healthy controls (C) and Parkinson's Disease (P) patients from those with postural instability and gait difficulty (G) subtype and NPH. CA and SA are significantly lower in G patients compared to C and P while EI is significantly higher. Automation of these geometric measures is relatively accurate for EI and SA but further improvements are needed for CA.
|
|
3672.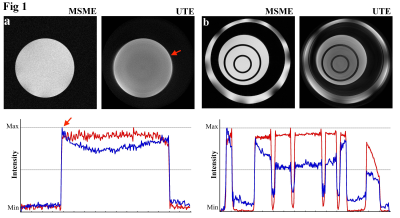 |
Post-acquisition image processing can reduce artifacts in UTE images
Yang Xia1, Farid Badar1, and Simon Miller1
1Physics, Oakland University, Rochester, MI, United States Poster Permission Withheld
Imperfect compensation for the eddy currents can cause artifacts in UTE images. Several post-acquisition processing methods can reduce artifacts in the images that are acquired in the radial k-space trajectories.
|
Session Topic: Software Tools and Image Analysis
Session Sub-Topic: Image Analysis 2
Digital Poster
Acquisition, Reconstruction & Analysis
3673.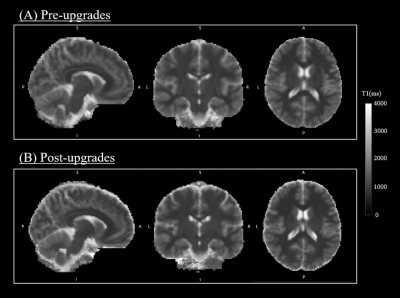 |
Evaluation of quantitative MRI parameters reproducibility across a major scanner upgrade: the example of T1
Ratthaporn Boonsuth1, Marco Battiston1, Francesco Grussu1,2, Marios C. Yiannakas1, Torben Schneider3, Rebecca S. Samson1, Ferran Prados1, and Claudia A. M. Gandini Wheeler-Kingshott1,4,5
1NMR research Unit, Queen Square MS Centre, Department of Neuroinflammation, UCL Queen Square Institute of Neurology, Faculty of Brain Sciences, University College London, London, United Kingdom, 2Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 3Philips Healthcare, Guildford, Surrey, United Kingdom, 4Brain MRI 3T Research Centre, IRCCS Mondino Foundation, Pavia, Italy, 5Department of Brain and Behavioural Sciences, University of Pavia, Pavia, Italy
Major MRI scanner upgrades are generally required to improve performance and image quality; however, they can also potentially introduce systematic changes in quantitative MRI (qMRI) metrics and affect their accuracy and precision. To date the evaluation of the effect of scanner upgrades has focussed mainly on volumetric measurements, whereas the effect on quantitative parametric maps remains unexplored, especially when comparing analog and digital signal pathways plus multiband. Here we report findings on quantitative T1 to assess the potential effect of scanner upgrades on qMRI. We found negligible differences, suggesting that T1 measurements remain stable following a major scanner upgrade.
|
|
3674.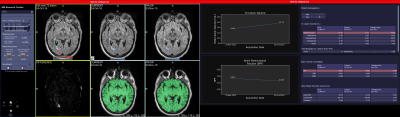 |
Quantitative MRI Metrics in Routine Clinical Practice: A Validation Study from a Large Heterogeneous Cohort of Multiple Sclerosis Patients
Adrian Tsang1, Mário J Fartaria2,3,4, Rodrigo D Perea1, Ricardo Corredor-Jerez2,3,4, Shirley Liao1, Tammie L.S. Benzinger5, Maria Laura Belfari1, Peter A Calabresi6, Carrie M Hersh7, Till Huelnhagen2,3,4, Stephen E Jones8, Hagen H Kitzler9, Nicholas Levitt1,
Yvonne W Lui10, Sara J Makaretz1, Robert Naismith5, Kunio Nakamura8, Dan Ontaneda8, Stephen Rao8, Alex Rovira11, Madalina E Tivarus12, James R Williams1, Richard A Rudick1, Tobias Kober2,3,4, and Elizabeth Fisher1
1Biogen, Cambridge, MA, United States, 2Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 3Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 4LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 5Washington University in St. Louis, St. Louis, MO, United States, 6Department of Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 7Cleveland Clinic Lou Ruvo Center for Brain Health, Cleveland, OH, United States, 8Cleveland Clinic, Cleveland, OH, United States, 9Center of Clinical Neuroscience, 'Carl Gustav Carus' University Hospital, Technische Universitaet, Dresden, Germany, 10New York University, New York, NY, United States, 11Vall d’Hebron University Hospital, Barcelona, Spain, 12University of Rochester Medical Center, Rochester, NY, United States
Use of MRI metrics for routine monitoring of MS patients has been hindered by lack of standardization, imprecise measurements, and workflow hurdles. To overcome these challenges, we developed a novel prototype, MSPie, as part of the MS PATHS initiative. The goal of this study was to assess the validity and suitability of MRI metrics automatically computed by MSPie in the MS clinical workflow.
|
|
3675.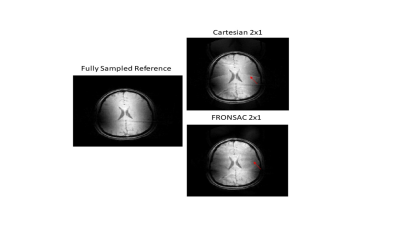 |
Experimental demonstration of 3D Cartesian-FRONSAC with new reconstruction approach
Yanitza Marie Rodriguez1, Gigi Galiana1, Enamul Buyian1, and Todd Constable1
1Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States
In this work we show the first experimental images, in both phantoms and humans, from 3D Cartesian Fast Rotary Nonlinear Spatial Acquisition (FRONSAC) imaging. FRONSAC adds oscillating nonlinear gradients to a standard Cartesian sequence, such as 3D GRE or MP-RAGE, both of which were acquired for this work. The additional modulation reduces undersampling artifacts and improves response to advanced reconstruction techniques, while maintaining other desirable image characteristics. In addition, we show that a PSF-based reconstruction, a generalization of the approach used in wave imaging, can be applied to FRONSAC data, yielding faster reconstructions that can be highly parallelized.
|
|
3676.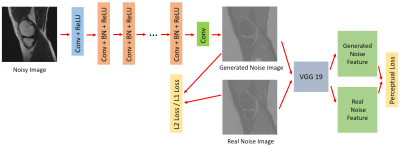 |
Perceptual Noise-Estimation based Method for MRI denoising with deep learning
Xiaorui Xu1, Siyue Li1, Shutian Zhao1, Chun Ki Franklin Au1, and Weitian Chen1
1CUHK lab of AI in radiology (CLAIR), Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, Hong Kong
Most of the methods in MRI denoising derive denoised images from corrupted images directly. DnCNN is a network that is used to remove Gaussian noise from natural images. The noise distribution in MRI images are often non-Gaussian due to the latest development of reconstruction algorithm and MRI hardware. In this work, we investigated the case when the noise follows Racian distribution. We utilized the idea of DnCNN and combine it with a perceptual architecture to remove Rician noise of MRI images. We demonstrated this method can generate ideal clean images.
|
|
3677.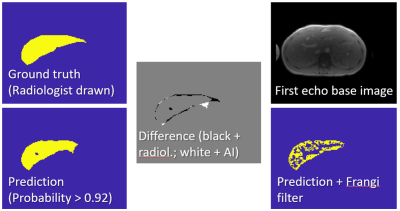 |
Automated MR HIC Determination using Deep Learning and Frangi Filters
Ralf Berthold Loeffler1,2, M. Beth McCarville2, Aaryani Tipirneni-Sajja2,3, Jane S Hankins4, and Claudia Maria Hillenbrand1,2
1Research Imaging NSW, University of New South Wales, Sydney, Australia, 2Diagnostic Imaging, St. Jude Children's Research Hospital, Memphis, TN, United States, 3Biomedical Engineering, University of Memphis, Memphis, TN, United States, 4Hematology, St. Jude Children's Research Hospital, Memphis, TN, United States
Hepatic iron content (HIC) quantification requires segmentation. Deep learning and Frangi Filtering allow to fully automate segmentation. 664 manually segmented data sets were available for training and testing a UNET. Data sets segmented by UNET were Frangi filtered for vessel exclusion, HIC was calculated using a published calibration, and correlated with HIC obtained with manual segmentation. Very good correlation (R2 > 0.99) with a correlation line close to unity was found. Fully automated HIC quantification using deep learning and Frangi filtering can lead to significant time savings in clinical practice.
|
|
3678.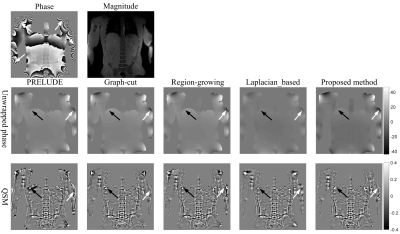 |
A Phase unwrapping Method by Phase Partition and Local Polynomial Fitting with Application to Abdominal Quantitative Susceptibility Mapping
Cheng Junying1, Xu Man1, Mei Yingjie2, Liu Liang1, Feng Yanqiu3, and Cheng Jingliang1
1Magnetic Resonance Department, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China, 2Philips Healthcare, Guangzhou, China, 3School of Biomedical Engineering, Southern Medical University, Guangzhou, China
The accurate recovery of underlying true phase is vital for a large number of applications, such as water/fat separation, quantitative susceptibility mapping and brain imaging. In this work, we propose a 3D phase unwrapping method based on phase partition and local polynomial fitting. The simulated and in vivo experiments demonstrate the proposed method can obtain perfect unwrapped results even in the regions with low SNR and rapidly changed phase, and can be applied to the abdominal QSM.
|
|
3679.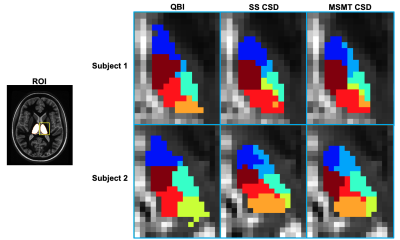 |
Fast, Automated DTI-based Thalamus Nuclei Segmentation
Charles Iglehart1, Adam Bernstein2, Ted Trouard3, Craig Weinkauf4, and Manoj Saranathan5
1Department of Electrical and Computer Engineering, University of Arizona, Tucson, AZ, United States, 2College of Medicine, University of Arizona, Tucson, AZ, United States, 3Department of Biomedical Engineering, University of Arizona, Tucson, AZ, United States, 4Department of Surgery, University of Arizona, Tucson, AZ, United States, 5Department of Medical Imaging, University of Arizona, Tucson, AZ, United States
Fast, accurate, and automatic thalamus segmentation is critical in evaluating the roles of individual thalamic nuclei in pathology and treatment. Many segmentation techniques developed to date involve the use of Diffusion Tensor MRI and are inhibited by their reliance upon i) time-consuming processing to produce an initial mask for the entire thalamus, and ii) orientation distribution functions incapable of modeling intricate small-scale fiber tract geometries. We present a technique that addresses these issues by i) greatly accelerating the masking process via template-based registration, and ii) using constrained spherical deconvolution to produce enhanced ODFs that drive a modified k-means clustering algorithm.
|
|
3680.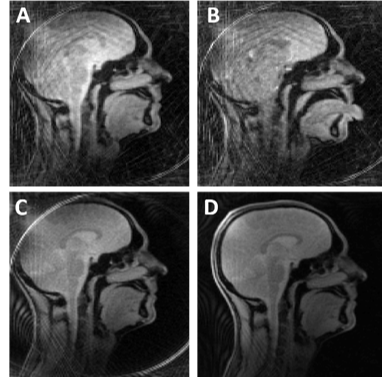 |
Real Time MRI at 70 frames per second: Establishing protocols for maxillofacial surgery applications.
Aneurin J Kennerley1, Isaac J Watson2, Lloyd A.E Bollans1, David A Mitchell3, and Angelika J Sebald1,3
1Chemistry, University of York, York, United Kingdom, 2Electronic Engineering, University of York, York, United Kingdom, 3York Cross Disciplinary Centre for Systems Analysis, University of York, York, United Kingdom
We showcase a 70+ frame per second Real Time MRI protocol that is robust and flexible for a number of different applications in maxillofacial surgery (including multi-plane monitoring speech and swallowing mechanisms) and associated follow up/long term monitoring for reducing depression and improving post-operative quality of life for patients. We explore the use of different food types to act as MR contrast agents to highlight changes in oral function. The project is co-created with a consulting Oral & Maxillofacial and Head & Neck Surgeon (Mr D Mitchell).
|
|
3681.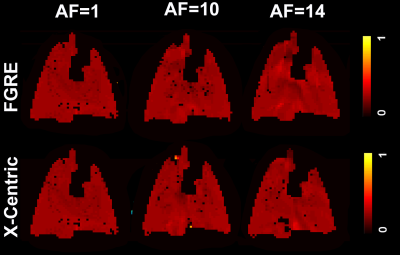 |
Feasibility of Dynamic Inhaled Gas MRI-based Measurements using Acceleration Factors of 10 and 14
Matthew S Fox1,2, Elise Woodward3, Marcus Couch4, Tao Li5, Iain Ball6, and Alexei V Ouriadov1,2
1Lawson Health Research Institute, London, ON, Canada, 2Physics and Astronomy, The University of Western Ontario, London, ON, Canada, 3The University of Western Ontario, London, ON, Canada, 4Montrel Neuro Institute, Montreal, QC, Canada, 5Thunder Bay Regional Research Institute,, Thunder Bay, ON, Canada, 6Philips Australia & New Zealand, North Ryde, Australia
We hypothesize that the SEM equation can be adapted for fitting the gas-density dependence of the MR signal similar to fitting time or b-value dependences S(n)=exp[-(nR)β], where 0<β<1, n is the image number and R is the apparent-fractional-ventilation parameter. This interpretation allows us to consider the signal-intensity variation as reflection of the underlying gas-density variation and hence, reconstruction of the under-sampled k-space using the adapted SEM equation. Lung fractional-ventilation maps have been generated using reconstructed images. We have demonstrated the feasibility of our approach using retrospective under-sampling mimicking acceleration factors of 10 and 14 in a small animal cohort.
|
|
3682.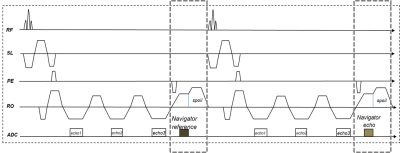 |
Self-Navigated MEDIC: an improved and reliable technique for cervical spine MRI
Qiong Zhang1, Yong Xiao Zhang2, Yuanyuan Kang3, and Wuyi Zhao1
1Siemens Shenzhen Magnetic Resonance, Ltd, Shen Zhen, China, 2MR Collaborations, Siemens Healthcare Ltd., Shenzhen, China, 3SW, Siemens Shenzhen Magnetic Resonance, Ltd, Shen Zhen, China
In this work, we developed a navigated Multi-Echo Data Imaging Combination (MEDIC) sequence for robust cervical spine (c-spine) imaging. In this sequence, a navigator echo is acquired in each repetition period to monitor the phase instability caused by respiration-induced field variations, and such instability among multiple echoes is subsequently compensated with a linear-phase evolution model. In vivo experimental results showed that the developed navigated MEDIC sequence outperformed the conventional MEDIC sequence and might be a potential technique for the diagnosis of cervical spine on 3T
|
|
3683.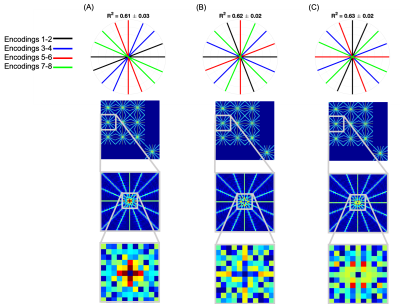 |
Radial sampling interactions in multi-dimensional sparse encoding problems using a joint decoding-reconstruction framework
Sophie Schauman1, Thomas W Okell1, and Mark Chiew1
1Wellcome Centre for Integrative Neuroimaging, NDCN, University of Oxford, Oxford, United Kingdom
Many physical properties cannot be directly measured with MRI, but are instead derived from a number of encoded measurements. Novel sampling methods in these regimes generally consider how to sample these encoded signals in k or k-t space, but not how to best sample across encodings. Here we present a study into how choice of encoding protocol interacts with the sampling and how these choices affect the multi-dimesnional point spread function. Simulations on vessel-encoded ASL angiography are used to study how these choices affect image reconstruction quality. We show that jointly decoding and reconstructing improves image reconstruction fidelity.
|
|
3684. |
Joint Compressed Sensing and Sensitivity Estimation for Free-Breathing Whole Heart CINE MRI Without ECG Gating
Jingyuan Lyu1, Jiali Zhong2, Yu Ding1, Qi Liu1, Lele Zhao3, Jian Xu1, Weiguo Zhang1, and Ruchen Peng2
1UIH America, Inc., Houston, TX, United States, 2Beijing LuHe Hospital, Capital Medical University, Beijing, China, 3United Imaging Healthcare, Shanghai, China
This abstract presents a new approach to compressed sensing cardiac MRI, which enables free-breathing whole heart coverage cine imaging within 30 seconds. Using a phased array coil, data was acquired continuously along Cartesian sampling trajectories using a lookup table without ECG gating. Each slice was continuously sampled for a fixed period of time, before the slice-selective RF excitation pulse switch to the next slice. In reconstruction, the approach jointly updates coil sensitivity maps and images, integrated with compressed sensing. In post-processing, virtual ECG is calculated based on unsupervised machine learning.
|
|
3685.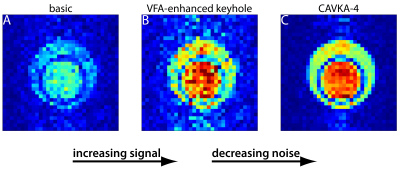 |
A Combined Approach of Variable Flip Angle, Keyhole and Averaging (CAVKA) for Accelerating the Acquisition of a low SNR Image Series
Hen Amit Morik1, Patrick Schuencke1, and Leif Schröder1
1Leibniz-Forschungsinstitut für Molekulare Pharmakologie (FMP), Berlin, Germany
We propose a method that is a Combined Approach of Variable Flip Angle (VFA), Keyhole undersampling and Averaging (CAVKA). It is designed to optimize the use of the limited magnetization and to accelerate the acquisition in MRI series that suffer from low SNR and thus require averaging. The method is applied to the acquisition of a CEST (chemical exchange saturation transfer) image series, where the sensed nucleus is hyperpolarized 129Xe. There it provides ~4-fold SNR increase compared to conventional imaging without averaging or 7-fold acceleration with the same SNR compared to imaging with averaging.
|
|
3686.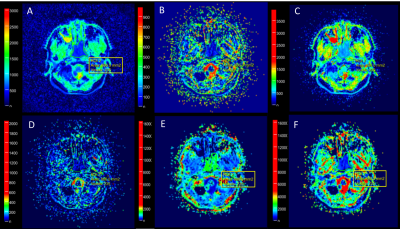 |
Partial Fourier Readout in Readout Segmentation of Long Variable Echo Trains-Based Imaging In Nasopharyngeal Carcinoma
Zilong Yuan1, Hao Chen1, Yaoyao He1, Huiting Zhang2, Xiaofang Guo1, Zhaoxi Zhang1, Yulin Liu1, and Robert Grimm3
1Radiology, Hubei Cancer Hospital, Tongji Medical College, Huazhong University of Science and Technology, wuhan, China, 2MR Scientific Marketing, Siemens Healthcare, Wuhan, China, 3MR Application Predevelopment,Siemens Healthcare, Erlangen, Germany
There is a potential influence of partial Fourier readout on readout segmentation of long variable echo trains (RESOLVE)-based intravoxel incoherent motion (IVIM) and diffusion kurtosis imaging (DKI) models. The aim of this study was to investigate the influence of partial Fourier readout on RESOLVE-based IVIM and DKI parameters in nasopharyngeal carcinoma. The results showed that partial Fourier readout has limited influence on DKI parameters, but significant influence on the perfusion parameters of IVIM.
|
|
3687.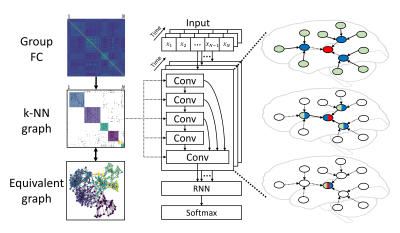 |
Connectivity-based Graph Convolutional Network for fMRI Data Analysis
Lebo Wang1, Kaiming Li2, and Xiaoping Hu1,2
1Department of Electrical and Computer Engineering, University of California, Riverside, Riverside, CA, United States, 2Department of Bioengineering, University of California, Riverside, Riverside, CA, United States
Graphs have been widely applied for ROI-based fMRI data analysis, in which the functional connectivity (FC) between all pairs of regions is thoroughly considered. Combined with convolutional neural networks, we define graphs based on FC and introduce a connectivity-based graph convolution network (cGCN) architecture for fMRI data analysis. cGCN allows us to extract spatial features within connectivity-based neighborhood for each frame and capture the temporal dynamics between frames. Our results indicate that cGCN outperforms traditional deep learning architectures on fMRI data analysis.
|
|
3688.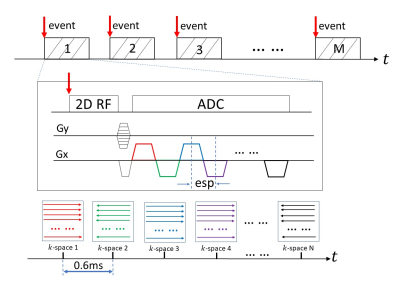 |
MRI with Sub-millisecond Temporal Resolution over a Reduced Field of View
Zheng Zhong1,2, Kaibao Sun2, Guangyu Dan1,2, Muge Karaman1,2, and Xiaohong Joe Zhou1,2,3,4
1Bioengineering, University of Illinois at Chicago, Chicago, IL, United States, 2CMRR, University of Illinois at Chicago, Chicago, IL, United States, 3Radiology, University of Illinois at Chicago, Chicago, IL, United States, 4Neurosurgery, University of Illinois at Chicago, Chicago, IL, United States
Sub-millisecond Periodic Event Encoded Dynamic Imaging or SPEEDI (also known as SMILE) has been reported to be capable of achieving sub-millisecond temporal resolution. However, the total scan times of this technique is typically very long. In this study, we have incorporated two techniques – reduced field of view (rFOV) and echo-train acquisition – into a SPEEDI sequence to substantially reduce the total scan times. This sequence, which we call re-SPEEDI, has been demonstrated in phantom experiments for capturing rapidly changing currents in a wire loop with a temporal resolution of 0.6ms-0.8ms.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance View the Poster
View the Poster Watch the Video
Watch the Video Back to Top
Back to Top