Scientific Session
Advances in Quantitative MRI
| Wednesday Parallel 5 Live Q&A | Wednesday, 12 August 2020, 14:30 - 15:15 UTC | Moderators: Rüdiger Stirnberg |
 |
0877.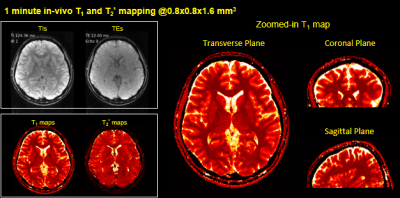 |
Fast Simultaneous T1, T2 and T2* Mapping at High Spatial Resolution using 3D Echo-planar Time-resolved Imaging (3D-EPTI)
Fuyixue Wang1,2, Zijing Dong1,3, Timothy G. Reese1, Lawrence L. Wald1,2, and Kawin Setsompop1,2
1A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Charlestown, MA, United States, 2Harvard-MIT Health Sciences and Technology, MIT, Cambridge, MA, United States, 33Department of Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States
An efficient quantitative mapping sequence based on 3D Echo-Planar Time-resolved Imaging (3D-EPTI) is proposed. The acquisition contains inversion recovery gradient echo readouts follow by GRASE-like readouts to provide sensitivity to T1, T2 and T2*. Fast k-TI-TE coverage is achieved by fusing highly-accelerated spatiotemporal CAIPI sampling with golden-angle radial-blade Cartesian under-sampling, where the reconstruction is performed using the subspace model. We demonstrate the high-efficiency of the proposed technique by obtaining multi-contrast images and quantitative maps at 1-mm isotropic resolution whole-brain in 3 minutes.
|
 |
0878.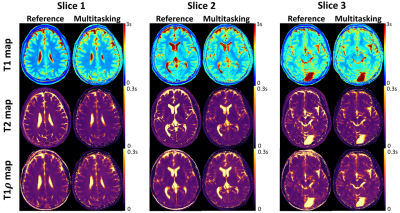 |
Motion-Resolved, 3D Whole-Brain Simultaneous T1, T2, and T1ρ Mapping using Multitasking with Application to Multiple Sclerosis: A Pilot Study
Sen Ma1,2, Anthony G. Christodoulou2, Nan Wang1,2, Marwa Kaisey3, Nancy L. Sicotte3, and Debiao Li1,2
1Department of Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 2Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 3Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, United States
Quantitative multi-parametric relaxometry MRI (e.g., T1, T2, and T1ρ mapping) can demonstrate longitudinal brain changes and enhance lesion contrasts against normal appearing matters in multiple sclerosis. Conventional methods that quantify these relaxation parameters are time-consuming and subject to motion, thus challenging for clinical practice. We present a novel approach that simultaneously quantifies T1, T2, and T1ρ with whole brain coverage in 9min, using the recently developed Multitasking framework that models the multidimensional image as a low-rank tensor. This technique is validated on healthy volunteers. We also demonstrate the feasibility of lesion characterization on relapsing remitting multiple sclerosis patients.
|
 |
0879.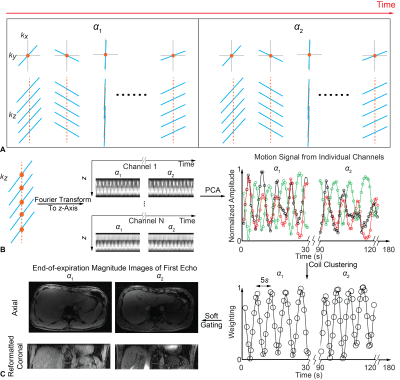 |
Free-Breathing Simultaneous Quantification of Liver T1, Fat and R2* with Variable Flip Angle Golden-Angle-Ordered 3D Stack-of-Radial MRI
Le Zhang1, Shu-Fu Shih1,2, Tess Armstrong1,3, and Holden H. Wu1,2,3
1Radiological Sciences, University of California, Los Angeles, Los Angeles, CA, United States, 2Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 3Physics and Biology in Medicine, University of California, Los Angeles, Los Angeles, CA, United States
Quantification of T1, proton-density fat fraction (PDFF), and R2* in the liver can provide information about a range of diseases. Existing Cartesian acquisition schemes generally require breath-holding, which limits spatial coverage and may be difficult for sick, elderly or pediatric patients. In this study, we propose a variable-flip-angle (VFA) golden-angle-ordered (GA) 3D stack-of-radial sequence that can provide multiparametric mapping with volumetric liver coverage in three minutes during free-breathing and with intrinsic motion compensation capability. Pilot studies in healthy subjects demonstrate agreement with reference breath-held scans and good measurement repeatability.
|
0880.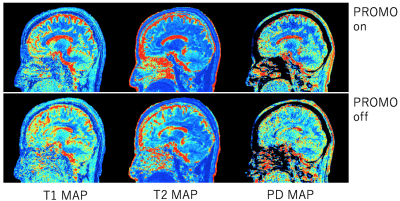 |
Prospective motion corrected 3D multi-parametric imaging
Naoyuki Takei1, David Shin2, Dan Rettmann3, Shohei Fujita4,5, Issei Fukunaga4, Akifumi Hagiwara4, Ken-Pin Hwang6, Marcel Warntjes7, Shigeki Aoki4, Suchandrima Banerjee2, and Hiroyuki Kabasawa1
1GE Healthcare, Tokyo, Japan, 2GE Healthcare, Menlo Park, CA, United States, 3GE Healthcare, Rochester, MN, United States, 4Juntendo University School of Medicine, Tokyo, Japan, 5The University of Tokyo Graduate School of Medicine, Tokyo, Japan, 6The University of Texas M.D. Anderson Cancer Center, Houston, TX, United States, 7SyntheticMR, Linkoping, Sweden
To aim for reliable parametric mapping to motion artifact, prospective motion correction was integrated to a multi-parametric technique, 3D QALAS. 2D Spiral navigators were inserted into wait times in the QALAS without impacting scan time for motion tracking and correction. The effectiveness of prospective motion correction was demonstrated. The proposed technique is expected to yield prospectively motion corrected 3D brain volumetric images of multiple contrasts and quantitative mappings.
|
|
0881.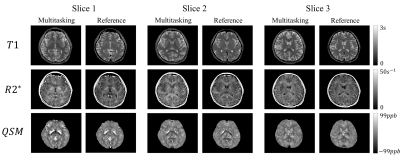 |
Simultaneous quantitative mapping of T1, R2* and susceptibility with magnetic resonance multitasking
Tianle Cao1,2, Nan Wang1,2, Sen Ma1,2, Yibin Xie1, Sara Gharabaghi3, E. Mark Haacke3,4,5, Anthony G. Christodoulou1, and Debiao Li1
1Biomedical Imaging Research Institute, Cedars Sinai Medical Center, Los Angeles, CA, United States, 2Bioengineering Department, University of California, Los Angeles, Los Angeles, CA, United States, 3Magnetic Resonance Innovations, Inc, Bingham Farms, MI, United States, 4Wayne State University School of Medicine, Detroit, MI, United States, 5The MRI Institute for Biomedical Research, Bingham Farms, MI, United States
A new approach for simultaneous quantitative mapping of T1, R2* and susceptibility was presented in this work. This technique employed IR pulses followed by N=152 segments of multi-echo FLASH readout. We were able to reconstruct the images for each echo time and inversion time under the multitasking framework for furhther analysis. Results of both visual comparison and statistical analysis showed that our proposed method agreed well with the reference but were more time efficient and robust to interscan motion.
|
|
0882.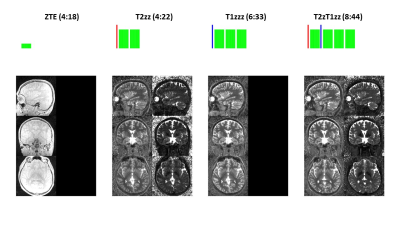 |
PSST … Parameter mapping Swift and SilenT
Florian Wiesinger1,2, Emil Ljungberg2, Mathias Engström3, Sandeep Kaushik1, Tobias Wood2, Steven Williams2, Gareth Barker2, and Ana Beatriz Solana1
1GE Healthcare, Munich, Germany, 2King’s College London, London, United Kingdom, 3GE Healthcare, Stockholm, Sweden
Here we describe a new framework for 3D, high-resolution Parameter mapping in a Swift and SilenT manner, termed PSST. The method combines T1 and T2 contrast preparation with segmented, silent, zero TE (ZTE) image encoding and an analytical signal model. Four canonical schemes are presented and demonstrated in phantom and in-vivo brain experiments.
|
|
 |
0883.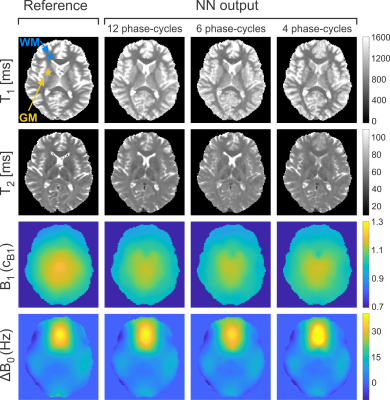 |
Deep-Learning Driven Acceleration of Multi-Parametric Quantitative Phase-Cycled bSSFP Imaging
Rahel Heule1, Jonas Bause1, Orso Pusterla2,3,4, and Klaus Scheffler1,5
1High Field Magnetic Resonance, Max Planck Institute for Biological Cybernetics, Tübingen, Germany, 2Institute for Biomedical Engineering, University and ETH Zurich, Zurich, Switzerland, 3Division of Radiological Physics, Department of Radiology, University Hospital Basel, Basel, Switzerland, 4Department of Biomedical Engineering, University of Basel, Basel, Switzerland, 5Department of Biomedical Magnetic Resonance, University of Tübingen, Tübingen, Germany
Prominent asymmetries in the bSSFP frequency profile in tissues with distinct fiber pathways are known to be a confounding factor in the quantification of relaxation times from a series of phase-cycled scans. It has been demonstrated that the resulting bias can be eliminated by training artificial neural networks using gold standard relaxation times as target. Here, the ability of neural networks to not only provide gold standard brain tissue T1 and T2 values as well as field map estimates (B1, ∆B0) but also to highly accelerate the acquisition by reducing the number of phase-cycles is explored.
|
0884.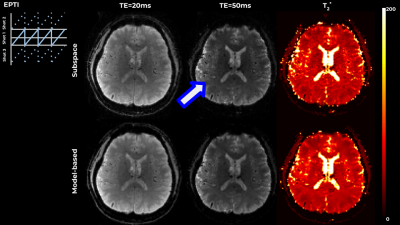 |
Flexible model-based reconstruction through generalized cycled parameter splitting approach.
Gilad Liberman1, Fuyixue Wang1, Zijing Dong1, and Kawin Setsompop1
1Department of Radiology, Massachusetts General Hospital, Charlestown, MA, United States
Model-based reconstruction approaches benefit from tight representation of the signal and from optimization on meaningful quantitative parameter maps, while requiring advanced algorithms and increased computational resources. We propose a generalized iterative thresholding algorithm with parameter splitting for model-based reconstruction, along with an efficient implementation. The approach is flexible and generalizable to problems in various MRI domains. We demonstrate it on the common image with phase evolution and signal decay model tackled with multi-echo GRE and Echo-Planar Time-resolved Imaging (EPTI), resulting in better image quality in comparison with GRAPPA and subspace constrained reconstruction, and increased z-scores in a low-SNR functional experiment.
|
|
0885.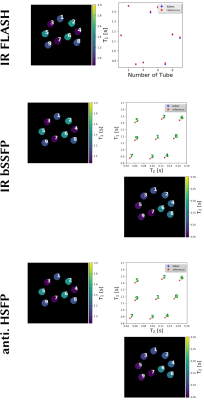 |
Generic Quantitative MRI using Model-Based Reconstruction with the Bloch Equations
Nick Scholand1,2, Xiaoqing Wang1,2, Sebastian Rosenzweig1,2, H. Christian M. Holme1,2, and Martin Uecker1,2
1Department of Interventional and Diagnostic Radiology, University Medical Center, Göttingen, Germany, 2German Centre for Cardiovascular Research, Göttingen, Germany Conventional quantitative MRI estimates parameters by fitting a known analytical signal model to pixels of images with different contrasts. By combining image reconstruction and the signal model into one non-linear inverse problem, model-based reconstruction methods can estimate the parameters directly from k-space. Avoiding the acquisition and reconstruction of intermediate images they require much less data. Furthermore, they can be directly combined with parallel imaging and compressed sensing, but still rely on analytical models and carefully designed MRI sequences. |
|
0886.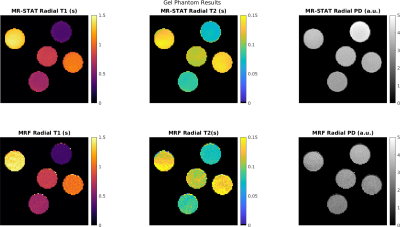 |
Extension of MR-STAT to non-Cartesian and gradient-spoiled sequences
Oscar van der Heide1,2, Alessandro Sbrizzi1,2, Tom Bruijnen1,3, and Cornelis van den Berg1,2
1Computational Imaging Group for MR Diagnostics and Therapy, Center for Image Sciences, University Medical Center Utrecht, Utrecht, Netherlands, 2Department of Radiology, Division of Imaging and Oncology, University Medical Center Utrecht, Utrecht, Netherlands, 3Department of Radiotherapy, Division of Imaging & Oncology, University Medical Center Utrecht, Utrecht, Netherlands
MR-STAT is a framework for obtaining multi-parametric quantitative MR maps using data from single short scans. A single large-scale optimization problem is solved in which spatial localisation of signal and estimation of tissue parameters are performed simultaneously. In previous work, MR-STAT was presented using gradient-balanced sequences with linear, Cartesian readouts. To demonstrate the generic nature of the MR-STAT framework and to explore potentially more efficient acquisition schemes, we extend MR-STAT to non-Cartesian gradient trajectories as well as gradient-spoiled sequences. We compare the our results from golden angle radial, gradient-spoiled acquisitions to low-rank ADMM MRF reconstructions on the same data sets.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Video
Watch the Video Back to Top
Back to Top