Oral - Power Pitch Session
MRS: New Developments, Applicatons, & Fighting the Noise
| Monday Parallel 5 Live Q&A | Monday, 10 August 2020, 15:15 - 16:00 UTC | Moderators: Masoumeh Dehghani & Ralf Mekle |
 |
0363.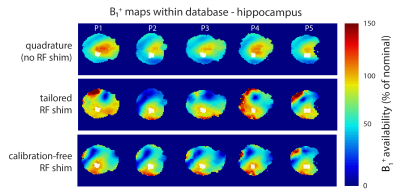 |
Calibration-free regional RF shims for localised MR spectroscopy
Adam Berrington1, Michal Povazan2, Christopher Mirfin1, Stephen Bawden1, Richard Bowtell1, and Penny Gowland1
1Sir Peter Mansfield Imaging Centre, School of Physics and Astronomy, University of Nottingham, Nottingham, United Kingdom, 2Russel H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University School Of Medicine, Baltimore, MD, United States
RF shimming can increase B1+ availability, which is critical for robust localised MR spectroscopy at ultra-high field. Shim calibration is performed on a region-wise basis and is, therefore, time consuming. Additionally, B1 distributions become difficult to predict. Recent work has shown that ‘universal’ pulses can be generated offline – avoiding the need for calibration. Here, we determine static calibration-free RF shims, optimised over 5 heads, for 3 different brain regions. B1+ availability using calibration-free shims was significantly higher than quadrature and comparable to tailored shimming. High quality spectra were also obtained from 3 regions with the calibration-free shims.
|
 |
0364.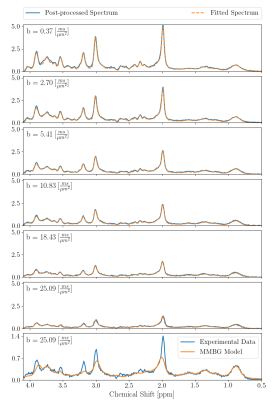 |
Diffusion-weighted MRS at short TE using a Connectom system: non-Gaussian metabolite diffusion and macromolecular signals in human brain
Kadir Şimşek1, André Döring1, André Pampel2, Harald E. Möller2, and Roland Kreis1
1Department of Radiology and Biomedical Research, University of Bern, Bern, Switzerland, 2Max-Planck Institute For Human Cognitive and Brain Sciences, Leipzig, Germany
Diffusion-weighted MRS was successfully implemented on a 3T Connectom system reaching b-values of 25 ms/um2 at a short TE of 30 ms and a moderate TM of 65 ms. Motion-compensation based on the water peak was found feasible up to the highest b-value and can be supplemented by scaling to the macromolecule peak intensity at 0.9 ppm. Non-Gaussian diffusion behavior was detected for multiple metabolites and was modeled with biexponential and kurtosis representations. In addition, a macromolecular spectrum could be determined by diffusion weighting and simultaneous modeling, which can now be used for quantification in clinical short TE MRS.
|
 |
0365.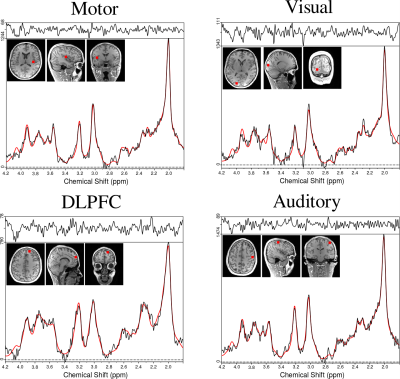 |
Intra-session and inter-subject variability of 3D-FID-MRSI using single-echo volumetric EPI navigators at 3T
Philipp Moser1, Korbinian Eckstein1, Lukas Hingerl1, Michael Weber1, Stanislav Motyka1, Bernhard Strasser1,2, Andre van der Kouwe3, Simon Robinson1, Siegfried Trattnig1,4, and Wolfgang Bogner1
1High Field MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 2Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Harvard Medical School, Massachusetts General Hospital, Vienna, MA, United States, 3Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology,, Harvard Medical School, Massachusetts General Hospital, Boston, MA, United States, 4Christian Doppler Laboratory for Clinical Molecular MR Imaging, Vienna, Austria
We demonstrate the combination of 3D free induction decay proton MR spectroscopic imaging and spatial encoding via concentric-ring trajectories at 3T. To improve the reliability a well as the temporal stability, single-echo, imaging-based volumetric navigators for real-time motion/shim-correction were additionally integrated. All intra-subject coefficients of variation and most of the inter-subject coefficients of variation obtained with motion/shim-correction were lower (i.e., better) than without and resulted in higher SNRs and lower CRLBs.
|
 |
0366.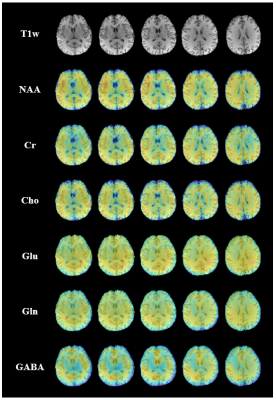 |
Rapid High-Resolution Mapping of Brain Metabolites and Neurotransmitters Using Hybrid FID/SE-J-Resolved Spectroscopic Signals
Yibo Zhao1,2, Yudu Li1,2, Jiahui Xiong1,2, Rong Guo1,2, Yao Li3,4, and Zhi-Pei Liang1,2
1Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 4Med-X Research Institute, Shanghai Jiao Tong University, Shanghai, China
J-resolved MRSI is a powerful tool for separating overlapping resonances in conventional MRSI, which is especially useful for mapping neurotransmitters like γ-aminobutyric acid and glutamate. A major practical limitation of J-resolved MRSI lies in its long data acquisition time required to sample the high-dimensional data space using spin-echo-based sequences. In this work, we present a novel hybrid FID/SE data acquisition scheme to accelerate J-resolved MRSI. The proposed method has been validated using phantom and in vivo experimental data, producing high-quality 3D spatial maps of brain metabolites and neurotransmitters within clinically feasible time.
|
 |
0367.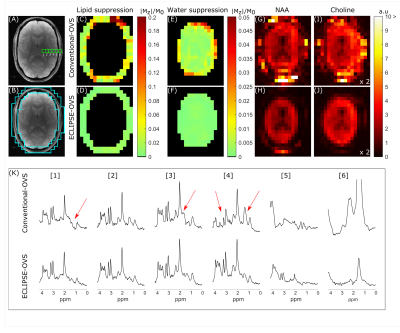 |
Robust Outer Volume Suppression Utilizing Elliptical Pulsed Second Order Fields (ECLIPSE) for Human Brain Proton MRSI
Chathura Kumaragamage1, Henk M De Feyter1, Peter B Brown1, Scott McIntyre1, Terence W Nixon1, and Robin A de Graaf1
1Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States
Extracranial lipid contaminants impede the reliable and accurate metabolite quantification with human brain MRSI. Elliptical localization with pulsed second order fields (ECLIPSE) was previously demonstrated for MRSI with inner volume selection (IVS), providing robust lipid suppression with improved elliptical brain coverage relative to a cubical ROI. In this work, alternative ECLIPSE-based OVS and IVS sequences were developed for human brain MRSI at 4T. Both ECLIPSE methods provide > 100-fold mean lipid suppression for short-TE MRSI. In addition, ECLIPSE-OVS consumes 30% of the power required by a traditional 8-slice OVS method, making ECLIPSE-OVS attractive for high field MRSI.
|
 |
0368.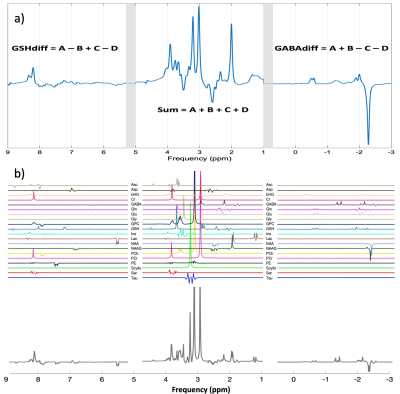 |
HERCULES and ConCAT: Simultaneous modelling and fitting of 11 metabolites using LCModel
Diana Rotaru1, Georg Oeltzschner2,3, Richard Edden3,4, and David Lythgoe1
1Neuroimaging, King's College London, London, United Kingdom, 2Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 4Radiology and Radiological Science, The Johns Hopkins University School of Medicine,, Baltimore, MD, United States
Advanced spectral editing techniques have focused mainly on GABA and GSH discrimination and detection. HERCULES, unlike all predecessor sequences, delivers a reliable detection method for GABA and GSH, but also for ascorbate, aspartate, 2-hydroxyglutarate, lactate, NAA and NAAG. However, current analysis methods have not been optimized for HERCULES analysis. We investigated simultaneous LCModel fitting of the concatenated sum and difference spectra calculated from HERCULES data. The concatenated (ConCAT) approach enables the use of all available spectral information. Compared to traditional single-spectrum analysis, ConCAT yielded improved results, with lower coefficients of variation obtained for concentration estimates and Cramér-Rao lower bounds.
|
 |
0369.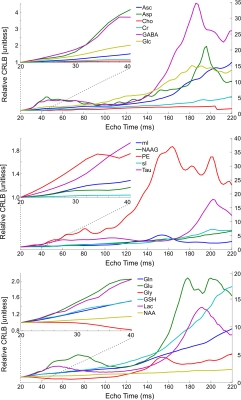 |
Optimization of echo time choice for seven common MRS pulse sequences through minimization of expected Cramér-Rao lower bounds.
Karl Landheer1 and Christoph Juchem1,2
1Biomedical Engineering, Columbia University, New York, NY, United States, 2Radiology, Columbia University, New York, NY, United States
It has recently been recommended to utilize the minimum echo time for non-editing magnetic resonance spectroscopic experiments. Despite this intuitive recommendation there is no comprehensive and systematic investigation into the choice of echo time across numerous sequences. Here the impact of echo time on the Cramér-Rao lower bounds for 17 different metabolites across the six most commonly used pulse sequences are investigated using simulated spectral shapes, as well as a MEGA-sLASER sequence for GABA quantification. Recommendations are provided for the choice of echo time which will minimize the expected Cramér-Rao lower bound for all metabolites and sequences in question.
|
0370.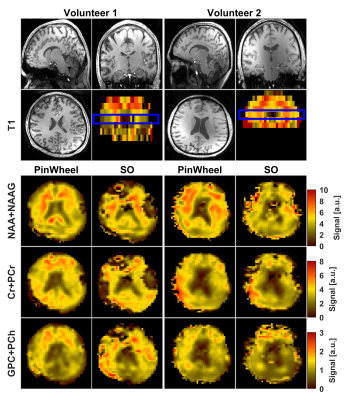 |
Whole-brain MR Spectroscopic Imaging with stack of Spirals Out-In k-space Trajectory at 7T
Morteza Esmaeili1,2, Bernhard Strasser1, Wolfgang Bogner3, Philipp Moser3, Zhe Wang4, and Ovidiu C. Andronesi1
1Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 2Department of Diagnostic Imaging, Akershus University Hospital, Lørenskog, Norway, 3High-field MR Center, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 4Siemens Medical Solutions, Charlestown, MA, United States
Metabolic imaging using magnetic resonance spectroscopic imaging (MRSI) provides important biomarkers for brain neurochemistry. We developed a spiral-out-in (SOI) trajectory for human whole-brain MRSI at 7T to take advantage of increased sensitivity and spectral separation at ultra-high field. We hypothesized that spectral-spatial SOI sampling will provide higher signal-to-noise ratio(SNR) compared to spiral-out (SO) sampling by increasing the efficiency of data collection. We acquired data from phantom and six healthy volunteers. Metabolic maps, SNR, Cramér-Rao-Lower-Bounds (CRLB) were evaluated between SO and SOI acquisitions. By more efficient data points collection per repetition time, SOI provided a significant improvement in SNR and CRLB.
|
|
 |
0371.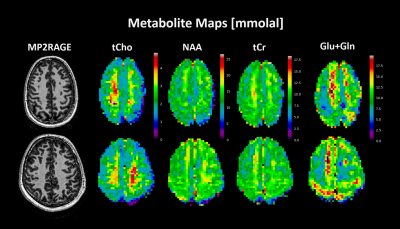 |
Quantitative Metabolite Mapping of the Human Brain at 9.4 T
Andrew Martin Wright1,2, Saipavitra Murali Manohar1,3, and Anke Henning1,4
1MRZ, Max Planck Institute for Biological Cybernetics, Tuebingen, Germany, 2International Max Planck Research School, University of Tuebingen, Tuebingen, Germany, 3University of Tuebingen, Tuebingen, Germany, 4Advanced Imaging Research Center, University of Texas Southwestern Medical Center, Dallas, TX, United States
Quantitative metabolite maps are reported from in vivo results at 9.4T. These maps are produced by quantifying with an internal water reference and utilize a novel T1 correction method applied to each voxel individually. Quantitative results allow cross-vendor and cross-site comparisons of results which may help to understand and characterize a variety neurological diseases.
|
0372.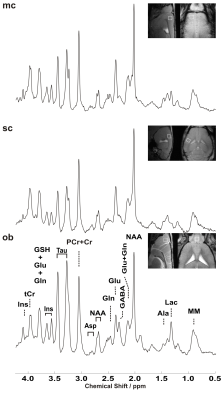 |
Towards sub-microlitre MRS in the mouse brain in vivo at ultra-high field
Alireza Abaei1, Dinesh K Deelchand2, Francesco Roselli3, and Volker Rasche1
1Core Facility Small Animal Imaging, Ulm University, Medical Center, ulm, Germany, 2Center for Magnetic Resonance Research, University of Minnesota, Minneapolis, MN, United States, 3German Center for Neurodegenerative Diseases (DZNE), Ulm, Germany
Several pathological conditions affect only a small volume of the cortex (such as the motor cortex in amyotrophic lateral sclerosis) and its characterization in mouse models is made impossible by the interference of normal, nearby cortical tissue. A sub-microlitre preclinical MRS technique was successfully implemented to detect subtle changes of the neurometabolite concentrations in three cortical areas. Employing LASER together with using cryogenically cooled RF coils significantly reduces the acquisition time to enable sub-microlitre MRS acquisition. Our findings demonstrate that neurochemical profiles of individual cortical brain regions can be reliably collected in pre-clinically feasible scan times.
|
|
0373.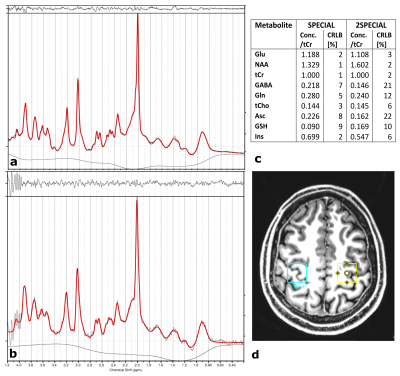 |
Adiabatic multiband inversion for simultaneous acquisition of 1H MR spectra from two voxels in-vivo at very short echo times
Layla Tabea Riemann1, Christoph Stefan Aigner1, Rüdiger Brühl1, Semiha Aydin1, Ralf Mekle2, Sebastian Schmitter1, Bernd Ittermann1, and Ariane Fillmer1
1Physikalisch-Technische Bundesanstalt (PTB), Braunschweig und Berlin, Germany, 2Center for Stroke Research Berlin, Charité-Universitätsmedizin, Berlin, Germany
In this work, a novel 1H MR spectroscopy sequence is proposed that provides the advantages of single voxel spectroscopy, such as high spectral bandwidth, a narrower point spread function, shorter measurement time and larger signal-to-noise-ratio, as compared to spectroscopic imaging while exciting more than one voxel. A multi-band adiabatic RF pulse was implemented into a SPECIAL sequence to simultaneously acquire the signal of two disjunct voxels at short echo times. The overlapping signal was decomposed using the SENSE algorithm. The new sequence was validated using a two-compartment phantom and its feasibility for in-vivo application is demonstrated at 7 T.
|
|
0374.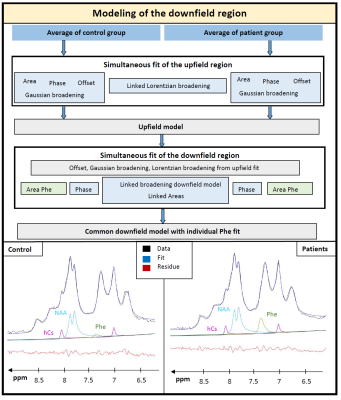 |
Quantification of phenylalanine with 1H MRS using optimized acquisition conditions and downfield background modeling
Maike Hoefemann1, Raphaela Muri2, Stephanie Abgottspon2, Johannes Slotboom3, Regula Everts2,4, Roman Trepp2, and Roland Kreis1
1Departments of Radiology and Biomedical Research, University of Bern, Bern, Switzerland, 2Department of Diabetes, Endocrinology, Clinical Nutrition and Metabolism, Bern University Hospital, University of Bern, Bern, Switzerland, 3Support Center for Advanced Neuroimaging, University Institute of Diagnostic and Interventional Neuroradiology Inselspital, Bern, Switzerland, 4Division of Neuropediatrics, Development & Rehabilitation, Pediatric University Hospital, University of Bern, Bern, Switzerland
For the quantification of the low-concentration metabolite phenylalanine (Phe) in patients with phenylketonuria using 1H magnetic resonance spectroscopy, optimal acquisition parameters and fitting procedures are crucial. Using a large voxel size and short TE helps to increase the signal-to-noise-ratio and allows restriction to a measurement time of 12min. Using the comparison of healthy controls vs. patients affords modeling of the unknown downfield region to develop a robust fitting model. Low CRLB of around 0.004mM proved the good precision of the quantification results, yielding cohort values of 0.019±0.01mM in controls and 0.142±0.02mM in patients.
|
|
 |
0375.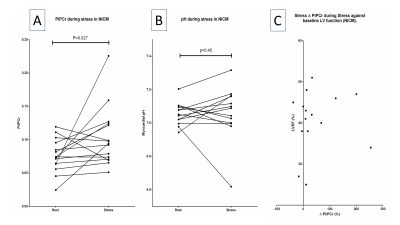 |
Myocardial Pi/PCr and pH during stress at 7T with STEAM 31P MRS in dilated cardiomyopathy; heart failure beyond the ejection fraction.
Andrew Apps1, Justin Lau1,2, Jane Ellis1, Mark Peterzan1, Moritz Hundertmark1, Damian Tyler3,4, Albrecht Ingo Schmid4,5, Stefan Neubauer6, Oliver Rider6, Ladislav Valkovic6,7, and Christopher T Rodgers8
1Cardiovascular Medicine, University of Oxford, Oxford, United Kingdom, 2Physiology, Anatomy and Genetics, University of Oxford, Oxford, United Kingdom, 3Physiology Anatomy and Genetics, University of Oxford, Oxford, United Kingdom, 4Oxford Centre for Magentic Resonance, University of Oxford, Oxford, United Kingdom, 5Medical University of Vienna, Vienna, Austria, 6Oxford Centre for Magnetic Resonance, University of Oxford, Oxford, United Kingdom, 7Imaging Methods, Slovak Academy of Sciences, Bratislava, Slovakia, 8Clinical Neurosciences, University of Cambridge, Cambridge, United Kingdom The addition of Pi/PCr quantification adds value over PCr/ATP for the characterisation of myocardial energetics. In defining the chemical shift of the Pi resonance, pH can also be computed. Such measurements however are hampered in 31P MRS due to the overlapping 2,3-DPG resonance. In harnessing the black blood contrast offered by STEAM, we successfully characterise Pi (and hence myocardial pH) in a cohort of patients with dilated cardiomyopathy. We go on to shown that in these patients (but not controls) Pi/PCr rises significantly during dobutamine stress, a finding that would significantly impair the free energy of ATP hydrolysis during exertion. |
0376.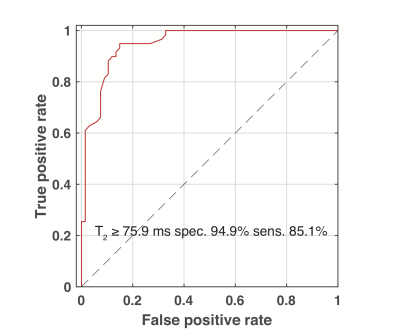 |
Fatty Acid Methylene T2 Can be Used to Separate Activatable Brown Adipose Tissue from Clavicular and Subcutaneous White Adipose Tissue in Humans
Ronald Ouwerkerk1, Jatin Raj Matta1, Ahmed Hamimi1, Aaron M Cypess2, Kong Y Chen3, and Ahmed Medhat Gharib1
1Biomedical and Metabolic Imaging Branch, NIDDK/NIH, Bethesda, MD, United States, 2Translational Physiology Section, Diabetes, Endocrinology, and Obesity Branch, NIDDK/NIH, Bethesda, MD, United States, 3Energy Metabolism Section, Diabetes, Endocrinology, and Obesity Branch, NIDDK/NIH, Bethedsa, MD, United States
Localized 1H-MRS was used to determine relaxation properties of fatty acid (FA) resonances in supraclavicular adipose tissue and distal white adipose tissue (WAT). Blinded to MRS results 18FDG-PETwas used to detect cold activated metabolism to identify active brown adipose tissue (BAT). Using the T2 of the FA methylene t a cutoff value of 76 ms this T2 can be used to distinguish BAT from distal or supraclavicular WAT with 85% sensitivity and 95% specificity
|
|
0377.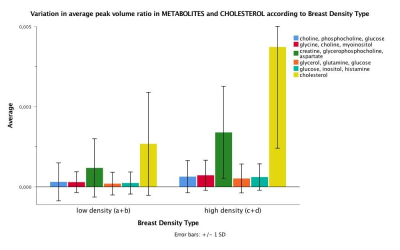 |
In vivo 2D COSY reveals metabolic and lipid variations between low and high BI RADS density breasts in an average breast cancer risk cohort of 65 women
Natali Naude1,2,3, Gorane Santamaria1,2,3, Thomas Lloyd3, Ian Bennett3, Jeremy Khoo3, Peter Malycha1,3, and Carolyn Mountford1,2,3
1Translational Research Institute, Brisbane, Australia, 2Queensland University of Technology, Brisbane, Australia, 3Princess Alexandra Hospital, Brisbane, Australia
Breast density is a strong risk factor for breast cancer with a four to six fold increase for those in BI-RADS high density group versus low density group. The current study acquired MRI and MRS in 65 women at average lifetime risk of developing breast cancer, and found statistically significant differences in various MR-visible lipids and metabolites as well as cholesterol between low and high breast density groups. Results implicate that increased metabolic activity underlies increased mammographic breast density. 2D COSY offers a non-invasive window into breast tissue chemistry, without the use of gadolinium-based contrast media.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Video
Watch the Video Back to Top
Back to Top