Oral - Power Pitch Session
Quantification, ML, and Tools
Session Topic: Quantification, ML, and Tools
Session Sub-Topic: Quantitative MRI: Reproducibility, Robustness & New Directions
Oral - Power Pitch
Acquisition, Reconstruction & Analysis
| Thursday Parallel 1 Live Q&A | Thursday, 13 August 2020, 14:20 - 15:05 UTC | Moderators: Gastao Cruz |
Session Number: PP-25
 |
1007.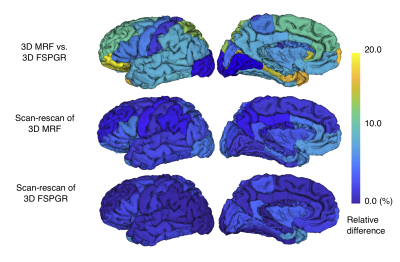 |
Reproducibility and Repeatability of Three-dimensional Magnetic Resonance Fingerprinting-based Human Brain Morphometry
Shohei Fujita1,2, Guido Buonincontri3,4, Matteo Cencini3,5, Naoyuki Takei6, Rolf F. Schulte7, Issei Fukunaga1, Akifumi Hagiwara1, Wataru Uchida1,8, Masaaki Hori9, Ryusuke Irie1,2, Koji Kamagata1, Osamu Abe2, and Shigeki Aoki1
1Department of Radiology, Juntendo University, Tokyo, Japan, 2Department of Radiology, The University of Tokyo, Tokyo, Japan, 3Imago7 Foundation, Pisa, Italy, 4IRCCS Stella Maris, Paris, Italy, 5Department of Physics, University of Pisa, Pisa, Italy, 6MR Applications and Workflow, GE Healthcare, Tokyo, Japan, 7GE Healthcare, Munich, Germany, 8Department of Radiological Sciences, Tokyo Metropolitan University, Tokyo, Japan, 9Department of Radiology, Toho University Omori Medical Center, Tokyo, Japan
Magnetic Resonance fingerprinting (MRF) provides simultaneous acquisition of T1 and T2 values with high reliability. However, the reproducibility and repeatability of human brain morphometry based on MRF still requires investigation. Here, we examined the feasibility of three-dimensional (3-D) MRF to evaluate the brain cortical thickness and volumetric analysis in healthy volunteers. Scan-rescan tests of both 3-D MRF and conventional 3D T1-weighted imaging were performed. For each sequence, the regional cortical thickness and volume of the subcortical structures were measured using automatic brain segmentation software. High agreement between conventional scans and scan-rescan repeatability in healthy human brains were observed.
|
 |
1008.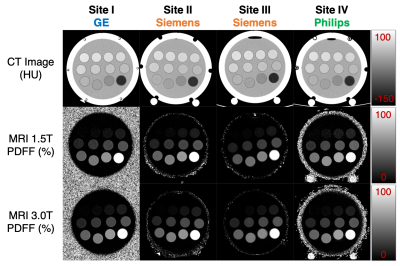 |
Multi-Site, Multi-Vendor Validation of the Accuracy and Reproducibility of Fat Quantification using a Novel MRI and CT Compatible Fat Phantom
Ruiyang Zhao1,2, Diego Hernando1,2, David T Harris1, Louis Hinshaw3, Ke Li1,2, Jessica Miller4, Perry J Pickhardt1, Ihab R Kamel5, Mahadevappa Mahesh5, Mounes Aliyari Ghasabeh5, Mustafa R Bashir6,7,8, Jean Shaffer6,7, Carolyn Lowry6,
Daniele Marin6, Takeshi Yokoo9, Lakshmi Ananthakrishnan9, Xinhui Duan9, and Scott B Reeder1,2,3,10,11
1Radiology, University of Wisconsin-Madison, Madison, WI, United States, 2Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 3Biomedical Engineering, University of Wisconsin-Madison, Madison, WI, United States, 4Human Oncology, University of Wisconsin-Madison, Madison, WI, United States, 5Radiology, John Hopkins University, Baltimore, MD, United States, 6Radiology, Duke University, Durham, NC, United States, 7Center for Advanced Magnetic Resonance Development, Duke University, Durham, NC, United States, 8Medicine, Duke University, Durham, NC, United States, 9Radiology, University of Texas Southwestern, Dallas, TX, United States, 10Medicine, University of Wisconsin-Madison, Madison, WI, United States, 11Emergency Medicine, University of Wisconsin-Madison, Madison, WI, United States
Accurate quantification of liver fat content is needed for early detection, staging, and treatment monitoring of non-alcoholic fatty liver disease. Chemical shift encoded MRI techniques enable accurate fat quantification though proton density fat fraction maps. CT is capable of quantifying fat based on the decrease in attenuation with increasing liver fat concentration. Current MR quantitative fat phantoms do not accurately mimic CT-based attenuation in the presence of liver fat. Therefore, the purpose of this work was to develop and validate the performance of a novel multimodality phantom that mimics the signals of liver fat in both MRI and CT.
|
1009.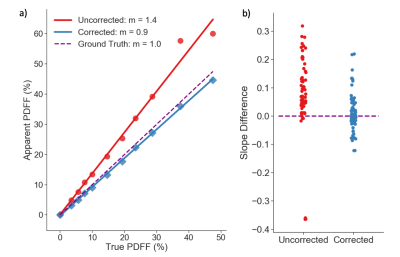 |
Multi-Center Phantom Validation of a Novel Method for Temperature Correction in PDFF Estimation using Magnitude Chemical Shift-Encoded MRI
Ruvini Navaratna1,2, Timothy J Colgan1, Ruiyang Zhao1,2, Houchun Harry Hu3, Mark Bydder4, Takeshi Yokoo5, Mustafa R Bashir6,7,8, Michael S Middleton9, Suraj D Serai10, Daria Malyarenko11, Thomas Chenevert11, Mark Smith3, Walter Henderson9,
Gavin Hamilton9, Yunhong Shu12, Claude B Sirlin9, Jean A Tkach13, Andrew T Trout13, Jean H Brittain14, Diego Hernando1,2, and Scott B Reeder1,2,15,16,17
1Radiology, University of Wisconsin-Madison, Madison, WI, United States, 2Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 3Radiology, Nationwide Children's Hospital, Columbus, OH, United States, 4Radiological Sciences, University of California - Los Angeles, Los Angeles, CA, United States, 5Radiology, University of Texas Southwestern Medical Center, Dallas, TX, United States, 6Radiology, Duke University Medical Center, Durham, NC, United States, 7Division of Gastroenterology, Duke University Medical Center, Durham, NC, United States, 8Center for Advanced Magnetic Resonance Development, Duke University Medical Center, Durham, NC, United States, 9Radiology, University of California - San Diego, San Diego, CA, United States, 10Radiology, Children's Hospital of Philadelphia, Philadelphia, PA, United States, 11Radiology, University of Michigan, Ann Arbor, MI, United States, 12Radiology, Mayo Clinic, Rochester, MN, United States, 13Radiology, Cincinnati Children's Hospital Medical Center, Cincinnati, OH, United States, 14Calimetrix, LLC, Madison, WI, United States, 15Biomedical Engineering, University of Wisconsin-Madison, Madison, WI, United States, 16Medicine, University of Wisconsin-Madison, Madison, WI, United States, 17Emergency Medicine, University of Wisconsin-Madison, Madison, WI, United States
Chemical shift-encoded MRI (CSE-MRI) is well-established to quantify proton density fat-fraction (PDFF) as a quantitative biomarker of hepatic steatosis.1 However, temperature is known to affect the accuracy and precision of PDFF quantification.2 In this study, we aim to characterize the effects of temperature on PDFF quantification using computer simulations, temperature-controlled phantom experiments, and a multi-center phantom study. Further, we present a novel method to minimize temperature-related fat quantification bias for magnitude-based CSE-MRI methods.
|
|
 |
1010.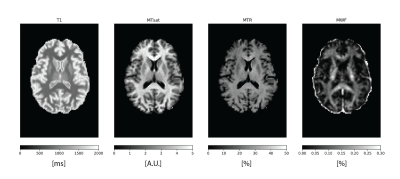 |
Myelin-sensitive Quantitative Maps: Two’s Company, Three’s a Crowd?
Matteo Mancini1,2,3, Eva Alonso-Ortiz2, Mara Cercignani1, Julien Cohen-Adad2, and Nikola Stikov2
1Department of Neuroscience, Brighton and Sussex Medical School, University of Sussex, Brighton, United Kingdom, 2NeuroPoly Lab, Polytechnique Montreal, Montreal, QC, Canada, 3CUBRIC, Cardiff University, Cardiff, United Kingdom
Are myelin-sensitive maps interchangeable? We performed a scan-rescan study in 5 subjects using T1 mapping, magnetization transfer and myelin water fraction. We found overall that all metrics have high scan-rescan repeatability and that they are highly correlated with each other. However, isolating the main factor behind this shared variance is not straightforward because of the different interplays between the measures. In the end, what will matter is to what extent these relationships are preserved in the presence of pathology.
|
1011.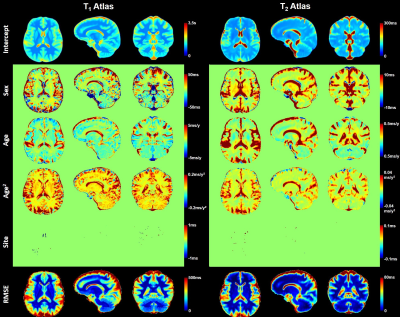 |
Large-scale quantitative atlases over the whole adult age range
Gian Franco Piredda1,2,3, Peipeng Liang4, Tom Hilbert1,2,3, Hongjian He5, Jean-Philippe Thiran2,3, Yi Sun6, Jianhui Zhong5,7, Kuncheng Li8,9, and Tobias Kober1,2,3
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3LTS5, École Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4School of Psychology, Capital Normal University, Beijing Key Laboratory of Learning and Cognition, Beijing, China, 5Center for Brain Imaging Science and Technology, Key Laboratory for Biomedical Engineering of Ministry of Education, College of Biomedical Engineering and Instrumental Science, Zhejiang University, Hangzhou, Zhejiang, China, 6MR Collaboration, Siemens Healthcare Ltd., Shanghai, China, 7Department of Imaging Sciences, University of Rochester, Rochester, NY, United States, 8Department of Radiology, Xuanwu Hospital, Capital Medical University, Beijing, China, 9Beijing Key Laboratory of Magnetic Resonance Imaging and Brain Informatics, Beijing, China
It was recently shown that brain atlases of normative relaxation times enable automated detection of tissue alterations on a single-subject basis. In this work, normative quantitative T1 and T2 atlases were obtained from a large-scale adult cohort of healthy volunteers (#997) covering a comprehensive age range (19-72y) in a multi-centric study including eleven sites. Atlases were derived by linearly modelling the inter-subject variability of T1/T2 while accounting for effects such as gender and age differences. Travelling subjects were scanned in nine centers with the same protocol, the comparison of the acquired maps showed good reproducibility of the employed relaxometry sequences.
|
|
1012.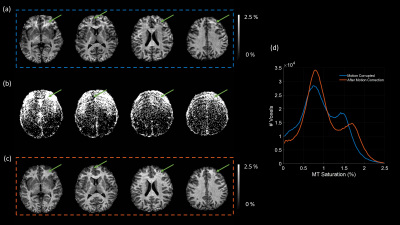 |
Intra-volume motion correction via Bayesian imputation in multi-parametric mapping (MPM) quantitative imaging
Mikael Brudfors1, Yaël Balbastre1, John Ashburner1, Siawoosh Mohammadi2,3, and Martina F Callaghan1
1Wellcome Centre for Human Neuroimaging, University College London, London, United Kingdom, 2Department of Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 3Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany
Intra-scan motion is a common source of artefacts in magnetic resonance imaging (MRI), which cannot be easily corrected. However, in quantitative MRI (qMRI), several volumes with varying parameters are acquired, and some sort of data redundancy exists. In this abstract, we propose a general framework where corrupted voxels are treated as missing entries and imputed using a Bayesian model of differently weighted MRI volumes. We demonstrate its efficacy in the context of various multi-parameter mapping (MPM) qMRI protocols, in which one volume is corrupted by motion. We show that the model can efficiently recover the corrupted data without introducing bias.
|
|
1013.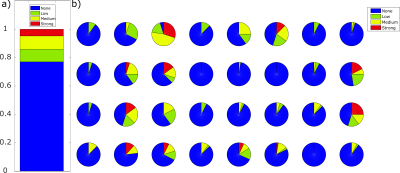 |
Data-driven Motion Detection for MR Fingerprinting
Gregor Körzdörfer1, Pedro Lima Cardoso2, Peter Bär2, Simone Kitzer2, Wolfgang Bogner2,3, Siegfried Trattnig2,3, and Mathias Nittka1
1Siemens Healthcare GmbH, Erlangen, Germany, 2High Field MR Centre, Department of Biomedical Imaging and Image-guided Therapy, Medical University of Vienna, Vienna, Austria, 3Christian Doppler Laboratory for Clinical Molecular MR Imaging, MOLIMA, Vienna, Austria
In contrast to qualitative MRI, motion artifacts can be more subtle in quantitative MRI methods such as Magnetic Resonance Fingerprinting (MRF). Errors caused by motion are not easily detectable by visual inspection of resulting maps. Hence, there is clear need for supporting the reliability of results with regard to motion-induced errors. We present a method to detect if significant through-plane motion occurred during an MRF scan, without external motion tracking devices or acquiring additional data. The method is based on classifying the spatiotemporal residuals either by eye or a neural network. The performance was successfully evaluated in a patient study.
|
|
 |
1014.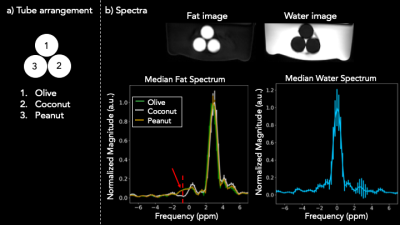 |
Free-breathing Abdominal Fat Spectroscopy with Multi-Echo Rosette k-space Sampling
Suma Anand1, Adam Michael Bush2, Christopher Michael Sandino3, Shreyas Vasanawala2, and Michael Lustig1
1Electrical Engineering and Computer Sciences, University of California, Berkeley, Berkeley, CA, United States, 2Radiology, Stanford University, Palo Alto, CA, United States, 3Electrical Engineering, Stanford University, Palo Alto, CA, United States
Obesity is a major cause of preventable morbidity and mortality in the US. A growing body of work suggests that triglyceride composition and its spatial distribution play a central role in this epidemic, necessitating the need for better non-invasive fat imaging. We propose a motion-robust acquisition scheme that combines the spatial resolution of MRI and the spectral resolution of MR spectroscopy using 2D multi-echo rosette k-space sampling. We validate the method with an oil phantom and demonstrate its motion robustness with a free-breathing in vivo acquisition.
|
1015.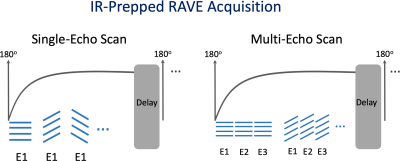 |
Stack-of-Stars Inversion-Recovery MRI for Free-Breathing T1 Mapping and IR-Prepared Fat/Water Separation
Li Feng1, Kai Tobias Block2, Thomas Benkert3, Ye Tian4, Chenyu Liu1, Fang Liu5,6, Zahi Fayad1, and Yang Yang1
1Biomedical Engineering and Imaging Institute and Department of Radiology, Icahn School of Medicine at Mount Sinai, New York, NY, United States, 2Center for Advanced Imaging Innovation and Research (CAI2R), Department of Radiology, New York University School of Medicine, New York, NY, United States, 3MR Applications Development, Siemens Healthcare GmbH, Erlangen, Germany, 4Department of Radiology and Imaging Sciences, University of Utah, Salt Lake City, UT, United States, 5Gordon Center for Medical Imaging, Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States, 6Department of Radiology, University of Wisconsin Madison, Madison, WI, United States
This work presents a framework for inversion-recovery (IR)-prepared stack-of-stars imaging and its applications for rapid free-breathing 3D liver MRI. Building upon a previously developed stack-of-stars 3D GRE sequence (RAVE: RAdial Volumetric Encoding), a non-selective 180o IR pulse has been implemented that is periodically played-out to achieve IR preparation (IR-Prepped RAVE). The new sequence allows (1) single-echo acquisition in combination with GRASP-Pro (imProved Golden-angle RAdial Sparse Parallel) reconstruction for free-breathing volumetric T1 mapping of the liver, and (2) multi-echo acquisition in combination with dynamic model-based reconstruction for IR-prepped and contrast-resolved fat/water separation.
|
|
 |
1016.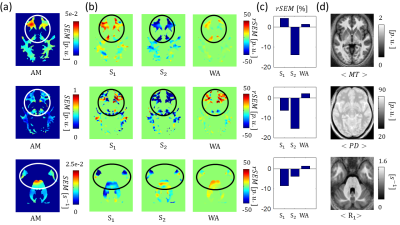 |
Identification and Correction of Errors in Quantitative Multi-Parameter Mapping (MPM)
Tobias Streubel1,2, Leonie Klock3, Martina Callaghan4, Simone Kühn3,5, Antoine Lutti6, Karsten Tabelow7, Nikolaus Weiskopf2, Gabriel Ziegler8,9, and Siawoosh Mohammadi1,2
1Institute for Systems Neuroscience, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 2Department of Neurophysics, Max Planck Institute for Human Cognitive and Brain Sciences, Leipzig, Germany, 3Department of Psychiatry and Psychotherapy, University Medical Center Hamburg-Eppendorf, Hamburg, Germany, 4Wellcome Trust Centre for Neuroimaging, UCL Institute of Neurology, UCL, London, United Kingdom, 5Center for Lifespan Psychology, Max Planck Institute for Human Development, Berlin, Germany, 6Laboratory for Research in Neuroimaging, Department of Clinical Neuroscience, Lausanne, Switzerland, 7Stochastic Algorithms and Nonparametric Statistics, Weierstrass Institute for Applied Analysis and Stochastics, Berlin, Germany, 8Institute of Cognitive Neurology and Dementia Research, Otto-von-Guericke-University, Magdeburg, Germany, 9German Center for Neurodegenerative Diseases, Magdeburg, Germany
We introduced novel error maps for proton density, longitudinal relaxation and magnetization transfer saturation rates that are more sensitive to artifacts than previously used error measures. We showed that they can be used to identify and down weigh local errors in the quantitative parameter maps for an experiment consisting of two successive multi-parameter mapping (MPM) measurements in a group of 10 healthy subjects.
|
1017.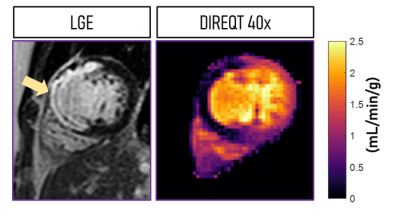 |
Model-based quantitative mapping for highly accelerated first-pass perfusion cardiac MRI
Teresa Correia1, Torben Schneider2, and Amedeo Chiribiri1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Philips Healthcare, Guildford, United Kingdom
First-pass perfusion cardiac MR (FP-CMR) is becoming essential for evaluating myocardial ischemia. However, FP-CMR requires ECG-gating and breath-holding, leading to a trade-off between spatial resolution and coverage. Moreover, perfusion abnormalities are often identified visually by highly trained operators. Recently, quantitative FP-CMR and compressed sensing (CS) have been proposed to reduce operator-dependency and moderately accelerate acquisitions, respectively. Here, a model-based reconstruction is proposed to directly estimate quantitative myocardial perfusion maps from highly undersampled acquisitions. Thus, allowing for higher spatial resolution and coverage than indirect methods, where dynamic images are reconstructed using CS and quantitative maps are obtained subsequently using tracer-kinetic modeling.
|
|
 |
1018.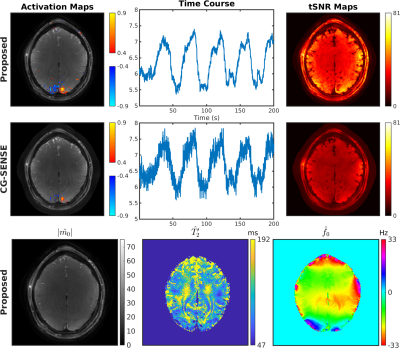 |
OSSI Manifold Model for High-Resolution fMRI Joint Reconstruction and Quantification
Shouchang Guo1, Douglas C. Noll2, and Jeffrey A. Fessler1
1Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States, 2Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Oscillating Steady-State Imaging (OSSI) is a new fMRI acquisition method that can provide high SNR signals, but does so at the expense of imaging time. We previously used a physics-based regularizer for high-quality, undersampled reconstruction by modeling the oscillating signal with physics parameters. However, the reconstructions were not quantitative, as the key parameter $$$T_2'$$$ for BOLD effects was not studied. In this work, to quantify MRI parameters of physiological importance, we jointly reconstruct the images and the parameters. The proposed manifold model reconstructs high-resolution images from 12-fold undersampled data, while also providing quantitative $$$T_2'$$$ estimates for fMRI.
|
1019.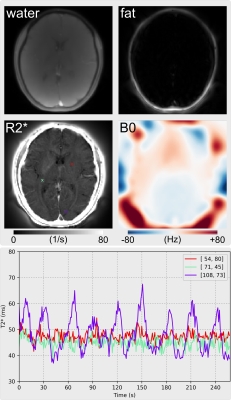 |
Dynamic Water, Fat, R2* and B0 Field Inhomogeneity Quantification Using Multi-Echo Multi-Spoke Radial FLASH
Zhengguo Tan1,2, Peter Dechent3, Xiaoqing Wang1,2, Nick Scholand1,2, Dirk Voit4, Jens Frahm2,4, and Martin Uecker1,2
1Diagnostic and Interventional Radiology, University Medical Center Göttingen, Göttingen, Germany, 2German Center for Cardiovascular Research (DZHK), Göttingen, Germany, 3Cognitive Neurology, University Medical Center Göttingen, Göttingen, Germany, 4Biomedizinische NMR, Max-Planck-Institute for Biophysical Chemistry, Göttingen, Germany
To achieve dynamic and simultaneous access to R2* relaxation rates, B0 field inhomogeneities and water/fat separation, we developed a model-based reconstruction technique on BART for continuous acquisitions based on undersampled multi-echo multi-spoke radial FLASH. Beside spatial smoothness constraints on coil sensitivity and B0 field maps, L1 wavelet regularization is applied to the water, fat and R2* maps. Preliminary results of brain fMRI data demonstrate significant T2* change from 37 to 67 ms in the occipital visual cortex. In addition, R2* mapping of free-breathing liver with only 15 RF shots per frame (194 ms temporal resolution) reveals increased R2* during inspiration.
|
|
 |
1020.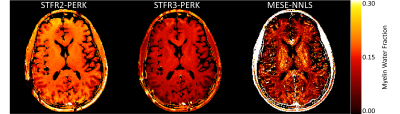 |
Myelin Water Imaging Using STFR with Exchange
Steven T. Whitaker1, Gopal Nataraj2, Jon-Fredrik Nielsen3, and Jeffrey A. Fessler1
1Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States, 2Medical Physics, Memorial Sloan Kettering Cancer Center, New York, NY, United States, 3Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States
Myelin water fraction (MWF) estimates are desirable for tracking the progression of demyelinating diseases such as multiple sclerosis. To address the long scan times of conventional MWF imaging methods, faster steady-state scans have been studied recently. One such steady-state scan is small-tip fast recovery (STFR). This work compares STFR-based MWF estimates using a two-compartment tissue model without exchange to those obtained using a three-compartment tissue model with exchange. Using a three-compartment model with exchange results in MWF estimates that are closer to traditional multi-echo spin echo (MESE) estimates.
|
1021.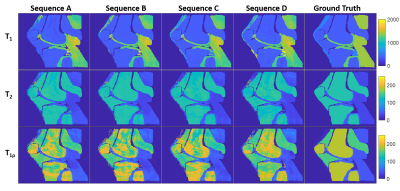 |
Quantification of T1ρ using magnetic resonance fingerprinting
Brendan Lee Eck1,2, Jeehun Kim2, Mingrui Yang2, and Xiaojuan Li2
1Cardiovascular and Metabolic Sciences, Cleveland Clinic, Cleveland, OH, United States, 2Biomedical Engineering, Cleveland Clinic, Cleveland, OH, United States
Quantitative T1ρ imaging has been studied for evaluating changes in tissue composition, in particular for detecting early cartilage degeneration in osteoarthritis. Magnetic resonance fingerprinting (MRF) provides a framework for rapid, robust acquisition of quantitative tissue property maps. Simulation experiments using spin-lock prepared MRF with different pulse schedules were conducted to demonstrate the feasibility of quantification of T1ρ in addition to T1 and T2. All tested sequences were sensitive to T1ρ and produced tissue property maps with major structures of interest. Differing accuracy and precision between sequences suggests opportunities for optimizing MRF for simultaneous T1, T2, and T1ρ quantification.
|

 Back to Program-at-a-Glance
Back to Program-at-a-Glance Watch the Video
Watch the Video Back to Top
Back to Top