Yuan Zheng1, Tao Feng1, Sirui Li2, Wenbo Sun2, Qing Wei3, Samo Lasic4, Danielle van Westen5, Karin Bryskhe4, Daniel Topgaard4,5, and Haibo Xu2
1UIH America, Houston, TX, United States, 2Zhongnan Hospital of Wuhan University, Wuhan, China, 3United Imaging Healthcare, Shanghai, China, 4Random Walk Imaging, Lund, Sweden, 5Lund University, Lund, Sweden
1UIH America, Houston, TX, United States, 2Zhongnan Hospital of Wuhan University, Wuhan, China, 3United Imaging Healthcare, Shanghai, China, 4Random Walk Imaging, Lund, Sweden, 5Lund University, Lund, Sweden
CNN was used to accelerate multidimensional dMRI, which characterizes µFA
with both directional and isotropic encodings and for the advanced versions requires longer scan and post-processing
times. High quality µFA maps were generated in real-time with only 50%
of the original encodings.
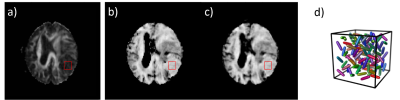
Figure 4: The
FA map of a meningioma case calculated using the conventional directional data
with a DTI model is shown in a). The µFA maps generated using
the Gamma model (80 encodings) and the CNN (40 encodings) are similar and shown
in b) and c) respectively. In the 9 × 9 ROI centered on the tumor, FA = 0.14 ±
0.04, µFA = 0.73 ± 0.05 and 0.70 ± 0.06 in b) and c). The low FA only indicates
there is no macroscopic anisotropy, while the high µFA reveals strong
microscopic anisotropy, as illustrated schematically in d).
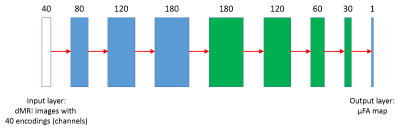
Figure 3: Structure of the CNN. The network has 4 convolution layers (blue) and 4
deconvolution layers (green). The number of channels is first expanded and then
gradually shrinked to 1, while the image dimension is kept at 112 × 112
throughout. Leaky-RELU is applied to all layers after convolution except the
output layer, which is followed by conventional RELU activation.