Fan Zhang1, Anna Breger2, Lipeng Ning1, Carl-Fredrik Westin1, Lauren J O'Donnell1, and Ofer Pasternak1
1Harvard Medical School, Boston, MA, United States, 2University of Vienna, Wien, Austria
1Harvard Medical School, Boston, MA, United States, 2University of Vienna, Wien, Austria
We obtain high quality and reliable brain tissue segmentation directly from diffusion MRI data by leveraging novel diffusion kurtosis imaging measures derived from the mean-kurtosis-curve method, and by applying a novel deep learning approach.
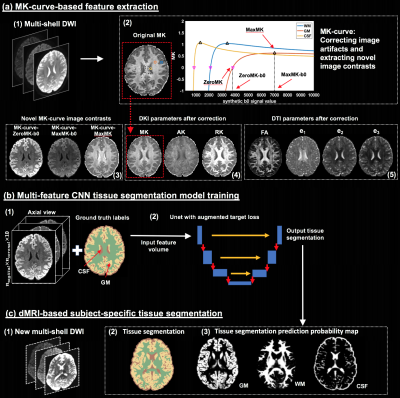
Figure-1: Method overview. Training is performed on high quality DWI where ground truth labels are obtained from T2w-based segmentation. From training DWI (a1), MK-curve (a2) is used to compute a set of 10 features: 3 novel MK-curve contrasts (a3), 3 DKI maps (a4) and 4 DTI maps (a5). Unet models (b) are trained using augmented target loss function (from axial, coronal and sagittal, respectively). The learned Unet is applied on a new DWI (c1), which could be of lower quality, to result with a predicted tissue segmentation, as well as segmentation prediction probability map per tissue type.
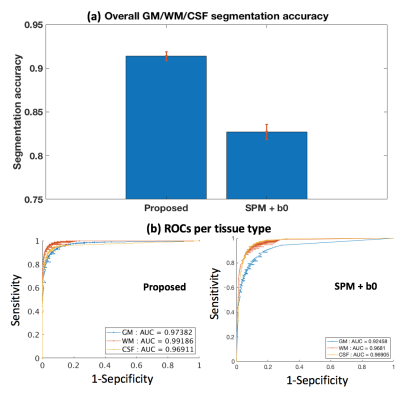
Figure-3: Quantitative comparison to the SPM+b0 method on test HCP data. (a) shows the mean and std of the segmentation accuracies across all test HCP subjects. (b) gives a receiver operating characteristic (ROC) analysis of the segmentation prediction probability map per tissue type, with respect to the SPM+T2w-based segmentation probability maps (Figure-4). Each data point on the ROC curve represents the comparison of binary segmentations, with the thresholds running from 0 and 1. Area under the curve (AUC) is reported for each method and each tissue type in the figure legend.
