Digital Posters
Modelling: Diffusion, Kinetics & More
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 15:00 - 16:00 |
2830.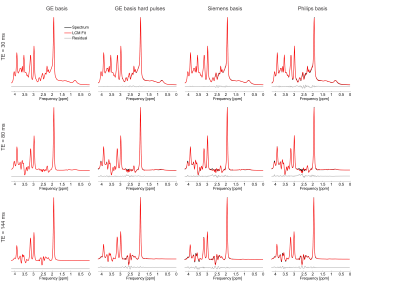 |
The effects of basis sets on magnetic resonance spectroscopy quantification for stock PRESS sequences, a simulation study
Karl Landheer1, Martin Gajdošík1, and Christoph Juchem1,2
1Biomedical Engineering, Columbia University, New York City, NY, United States, 2Radiology, Columbia University, New York City, NY, United States Realistic synthetic PRESS spectra were generated for three different echo times for each of the three major vendors. These spectra were then fit to the matched basis set (i.e., the basis set used to generate it), as well as the mismatched basis sets at the same echo time from other vendors, and the matched basis set but with the hard pulse approximation, to investigate how sensitive resulting quantification is to basis sets. It was found that the concentration for low-concentration metabolites is highly susceptible to small changes in basis sets (e.g., GABA varied by 115 ± 188%). |
|||
2831.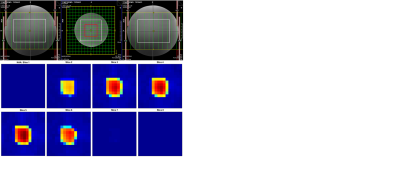 |
Reproducibility of Semi-LASER Localized Correlated Spectroscopic Imaging Using Concentric Ring Echo-Planar Trajectories
Andres Saucedo1, Manoj Kumar Sarma1, Uzay Emir2, James Sayre1, Paul M Macey3, and Michael Albert Thomas1
1Radiological Sciences, UCLA Geffen School of Medicine, Los Angeles, CA, United States, 2School of Health Sciences, Purdue University, West Lafayette, IN, United States, 3UCLA School of Nursing, Los Angeles, CA, United States Five-dimensional correlated spectroscopic imaging using concentric ring trajectories (5D COSI-CONCEPT) has been implemented. Since the maximum slew rates required for the concentric ring trajectories are lower than those for Cartesian echo-planar spectroscopic imaging for a given spectral bandwidth and spatial resolution, the eddy current issues are less challenging. The acquired 5D COSI-CONCEPT data was regridded along kx-ky and the accelerated kz-t1 data was reconstructed using group sparsity. Reproducibility of the sequence was evaluated in a brain phantom using 3T Prisma and Skyra MRI scanners. Pilot findings showed excellent coefficients of variance and intra-class correlation coefficients for all the six metabolites.
|
|||
2832.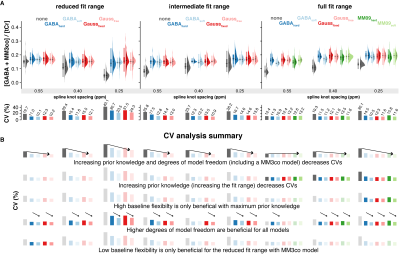 |
Linear-combination modeling of GABA-edited MEGA-PRESS at 3T: Evaluating different modeling strategies
Helge Jörn Zöllner1,2, Sofie Tapper1,2, Steve C. N. Hui1,2, Peter B. Barker1,2, Richard A. E. Edden1,2, and Georg Oeltzschner1,2
1Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States
Recent consensus states that linear-combination modeling (LCM) is the preferred method for modeling GABA-edited spectra. However, there is little peer-reviewed literature investigating how this should be performed. Here, we compare various modeling strategies for GABA-edited spectra, with different approaches to macromolecular signal parametrization, baseline stiffness, and fitting range. Variance of GABA+/tCr estimates decreased when prior knowledge was maximized, i.e., when a full fit range was used with a dedicated co-edited MM basis function. These new modeling strategies have been implemented in the MATLAB-based open-source MRS analysis toolbox Osprey (github.com/schorschinho/osprey).
|
|||
2833.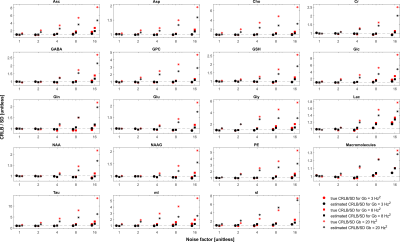 |
Are Cramér-Rao Lower Bounds an Accurate Estimate for Standard Deviations in Magnetic Resonance Spectroscopy?
Karl Landheer1 and Christoph Juchem1,2
1Biomedical Engineering, Columbia University, New York City, NY, United States, 2Radiology, Columbia University, New York City, NY, United States
Cramér-Rao Lower Bounds (CRLBs) have become the routine method to approximate standard deviations for magnetic resonance spectroscopy. CRLBs are theoretically a lower bound on the standard deviation. Realistic synthetic 3 Tesla spectra were used to investigate the relationship between estimated CRLBs, true CRLBs and standard deviations. It was demonstrated that although the CRLBs are theoretically truly a lower bound on the standard deviation this approximation is valid only as long as the model properly characterizes the data. In the case when the basis set deviates from the measured data it was shown that the CRLBs deviate substantially from standard deviations.
|
|||
2834.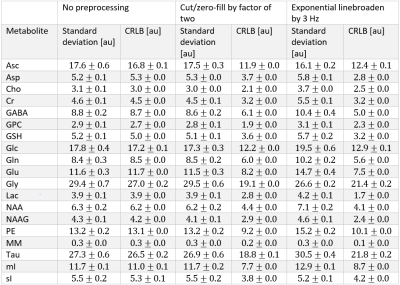 |
The effects of cutting/zero-filling and linebroadening on quantification of magnetic resonance spectra via maximum-likelihood estimation
Karl Landheer1 and Christoph Juchem1,2
1Biomedical Engineering, Columbia University, New York City, NY, United States, 2Radiology, Columbia University, New York City, NY, United States
It has recently been recommended that typical preprocessing tools, such as linebroadening, zero-filling and apodization (cutting), generally be avoided prior to signal quantification via consensus. To date, little explanation has been provided against these tools which have become commonplace. Here we demonstrate via realistic Monte Carlo simulations that such preprocessing tools may reduce the precision of the extracted parameters and artificially reduce the Cramér-Rao Lower Bounds and provide a theoretical outline for why they should be avoided.
|
|||
2835.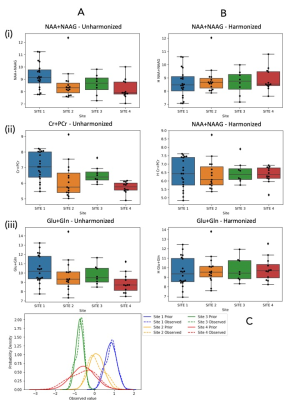 |
ComBat Empirical Bayes Model Supersedes Naive Methods for Statistical Harmonization of Multi-Site 1H MR Spectroscopy
Lasya P Sreepada1,2, Sam H Jiang1, Huijun Vicky Liao1, Katherine M Breedlove1, Eduardo Coello1, and Alexander P Lin1
1Radiology, Center for Clinical Spectroscopy, Brigham and Women's Hospital and Harvard Medical School, Boston, MA, United States, 2Radiology, Center for Biomedical Image Computing and Analytics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States
As medical imaging enters the information era, there is a rapidly increasing need for big data analytics. Robust pooling and harmonization of multi-site data across diverse cohorts is critical. We compare the performances of current basic methods with ComBat, an Empirical Bayes method that removes batch effects, in harmonizing 1H Brain MR Spectroscopy (MRS) of healthy controls from 4 sites. Basic harmonization did not bring metabolite means closer together and increased variance, while ComBat successfully removed significant site effects as determined by ANOVA and Levene's tests. These results may improve reproducibility and generalizability of MRS studies, especially in clinical space.
|
|||
2836.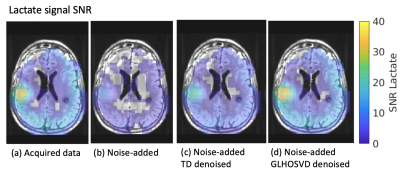 |
Assessment of Higher-Order SVD Rank Reduction Denoising on Dynamic Hyperpolarized [13C]pyruvate Metabolic Imaging Data on Patients with Glioma
Sana Vaziri1, Adam Autry1, Yaewon Kim1, Hsin-Yu Chen1, Jeremy W Gordon1, Marisa LaFontaine1, Jasmine Graham1, Janine Lupo1, Jennifer Clarke2, Javier Villanueva-Meyer1, Nancy Ann Oberheim Bush3, Duan Xu1, Susan M Chang2, Peder
EZ Larson1, Daniel B Vigneron1,4, and Yan Li1
1Department of Radiology and Biomedical Imaging, University of California, San Francisco, San Francisco, CA, United States, 2Department of Neurological Surgery, University of California, San Francisco, San Francisco, CA, United States, 3Department of Neurology, University of California, San Francisco, San Francisco, CA, United States, 4Department of Bioengineering and Therapeutic Science, University of California, San Francisco, San Francisco, CA, United States
Real-time monitoring of enzymatic conversion of [1-13C]pyruvate to [1-13C]lactate and [1-13C]bicarbonate in the brain can be performed using dynamic hyperpolarized 13C metabolic imaging. However, signal-to-noise ratios of bicarbonate are significantly lower than lactate and pyruvate, making it difficult to assess metabolic flux, particularly in lesions. Denoising techniques employing rank reduction via the multidimensional extension of singular value decomposition have recently been introduced for conventional MR images, diffusion-weighted images and HP-13C metabolic imaging. Here, we investigate the use of two higher-order singular value decomposition denoising techniques on dynamic hyperpolarized 13C metabolic images acquired from patients with glioma.
|
|||
2837. |
A Bayesian Approach for T2* Mapping with Built-in Parameter Estimation
Shuai Huang1, James J. Lah1, Jason W. Allen1, and Deqiang Qiu1
1Emory University, Atlanta, GA, United States
We propose a Bayesian approach with built-in parameter estimation to perform T2* mapping from undersampled k-space measurements. Compared to conventional regularization-based approaches that require manual parameter tuning, the proposed approach treats the parameter as random variables and jointly recovers them with T2* map. Additionally, the estimated parameters are adaptive to each dataset, this allows us to achieve better performances than regularization-based approaches where the parameters are fixed after the tuning process. Experiments show that our approach outperforms the state-of-the-art l1-norm minimization approach, especially in the low-sampling-rate regime.
|
|||
2838.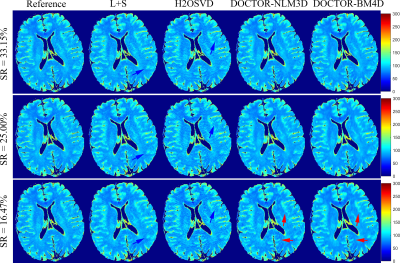 |
Denoising Induced Iterative Reconstruction for Fast $$$T_{1\rho}$$$ Parameter Mapping
Qingyong Zhu1, Yuanyuan Liu2, Zhuo-Xu Cui1, Ziwen Ke1, and Dong Liang1,2
1Research Center for Medical AI, SIAT, Chinese Academy of Sciences, Shenzhen, China, 2Paul C. Lauterbur Research Center for Biomedical Imaging, SIAT, Chinese Academy of Sciences, Shenzhen, China
We propose a novel DenOising induCed iTerative recOnstRuction framework (DOCTOR) to realize fast $$$T_{1\rho}$$$ parameter mapping from under-sampled k-space measurements. The proposed formulation constrains simultaneously intensity-based and orientation-based similarity between the reconstructed images and denoised prior images. Two state-of-art 3D denoising technologies are utilized including NLM3D and BM4D. The reconstruction alternates between two steps of denoising and a quadratic programming attacked by non-linear conjugate gradient method. The parameter maps are created from the reconstructed images using conventional fitting with an established relaxometry model. Through experiments in-vivo $$$T_{1\rho}$$$-weighted brain MRI datasets, we can observe superior image-quality of the proposed DOCTOR scheme.
|
|||
2839.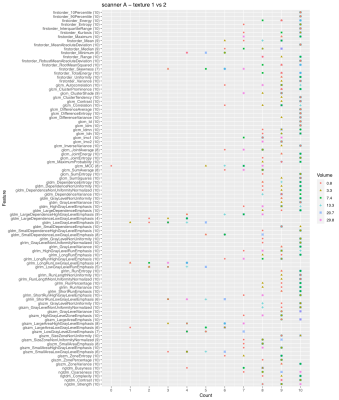 |
Discrimination of tumor texture based on MRI radiomic features: is there a volume threshold? A phantom study.
Linda Bianchini1, João Santinha2,3, Francesca Botta4, Daniela Origgi4, Marta Cremonesi4, and Alessandro Lascialfari1
1University of Pavia, Pavia, Italy, 2Champalimaud Center for the Unknown, Lisbon, Portugal, 3Instituto Superior Técnico, Lisbon, Portugal, 4European Institute of Oncology IRCCS, Milan, Italy
This study tested the hypothesis that some MRI-based radiomic features could lose their texture descriptive power below a volume threshold, compromising the robustness of prediction models. The ability of features to discriminate different textures as a function of the tumor volume was investigated on T2-weighted images of a customized pelvic phantom. The images were acquired on three scanners and considered three different textures, from a finer to a coarser one. The texture discriminative ability was shown to depend on tumor volume, with most features losing this property with a VOI smaller than 1 cm3 at 1.5 T.
|
|||
2840.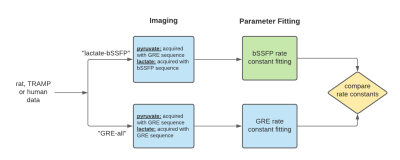 |
Fitting kinetic rate constants in metabolite-specific bSSFP hyperpolarized [1-13C]pyruvate MRI
Sule Sahin1,2, Shuyu Tang3, Manushka Vaidya2, and Peder E.Z. Larson2
1Graduate Program in Bioengineering, University of California, Berkeley and University of California, San Francisco, Berkeley, CA, United States, 2Radiology, University of California, San Francisco, San Francisco, CA, United States, 3HeartVista, Inc., Los Altos, CA, United States
An alternate to AUC ratio, fitting pyruvate to lactate rate constants (kPL) can be a powerful tool for quantification of hyperpolarized [1-13C]pyruvate studies. In this work, a model was developed to fit kPL values to a novel acquisition method where lactate was acquired with a stack-of-spiral bSSFP sequence. The model was utilized to fit kPL on three sets of in vivo data: healthy rat kidneys, mouse prostate tumors and human kidney tumors. It was shown that the fit kPL values matched those fit using an established GRE fitting method for complimentary GRE-acquired data sets.
|
|||
2841.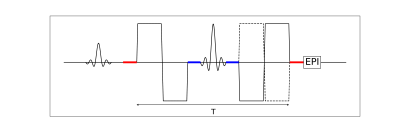 |
Time dependence of flow compensated intravoxel incoherent motion in tumor
Oscar Jalnefjord1,2, Louise Rosenqvist1, Mikael Montelius1, Lukas Lundholm1, Eva Forssell-Aronsson1,2, and Maria Ljungberg1,2
1Department of Radiation Physics, University of Gothenburg, Gothenburg, Sweden, 2Department of Medical Physics and Biomedical Engineering, Sahlgrenska University Hospital, Gothenburg, Sweden
This study provides initial results on the encoding-time dependence of IVIM parameters in tumor obtained from a combination of flow-compensated and non-flow-compensated diffusion-encoded data. This was made possible by constructing a pulse sequence capable of performing flow-compensated diffusion encoding with variable encoding time, which was validated through phantom measurements.
|
|||
2842.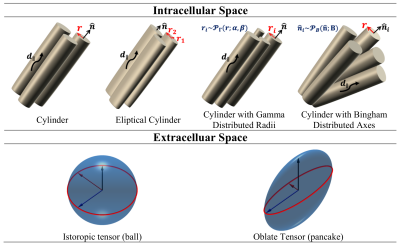 |
Comparison of compartmental models of diffusion MRI for assessing myocardial microstructure
Mohsen Farzi1, Irvin Teh1, Darryl McClymont2, Hannah Whittington2, Craig A. Lygate2, and Jürgen E. Schneider1
1Cardiovascular & Metabolic Medicine, University of Leeds, Leeds, United Kingdom, 2Cardiovascular Medicine, University of Oxford, Oxford, United Kingdom
Diffusion tensor imaging (DTI) is a valuable technique for interrogating tissue microstructure, but the estimated parameters remain an indirect characterisation of the underlying tissue architecture. For direct measurement of biophysical parameters, we propose a two-compartment model to quantify cardiomyocyte radius, volume fraction, and dispersion. The intra- and extra-cellular space were modelled using a cylinder with Bingham distributed axes and an oblate tensor. The model reduced root mean squared error by 5% compared to DTI, with volume fraction = 60%, radius = 5.8𝜇m, and dispersion in the sheetlet plane = 9°. These parameters could serve as biomarkers for characterisation of cardiomyopathies.
|
|||
2843.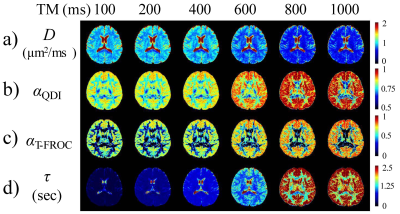 |
Time Dependency of the Continuous-Time Random-Walk Diffusion Model at Long Diffusion Times in the Human Brain
Guangyu Dan1,2, Yuxin Zhang3,4, Zheng Zhong1,2, Kaibao Sun1, Muge Karaman1,2, Diego Hernando3,4, and Xiaohong Joe Zhou1,2,5
1Center for MR Research, University of Illinois at Chicago, Chicago, IL, United States, 2Department of Bioengineering, University of Illinois at Chicago, Chicago, IL, United States, 3Department of Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 4Department of Radiology, School of Medicine and Public Health, University of Wisconsin-Madison, Madison, WI, United States, 5Departments of Radiology and Neurosurgery, University of Illinois at Chicago, Chicago, IL, United States
It has been increasingly reported that the diffusion-weighted MRI signal depends on not only the b-value but also the diffusion time. Investigations of diffusion model parameters on diffusion time can provide information on interaction between water molecules and their environment, thus helping reveal tissue microstructures. In this study, we focused on two special cases of a continuous-time random-walk (CTRW) diffusion model; and investigated the diffusion-time dependency of the CTRW parameters in the human brain. Our results showed significant dependency of the CTRW parameters on diffusion times in the range of 100-1000 ms.
|
|||
2844.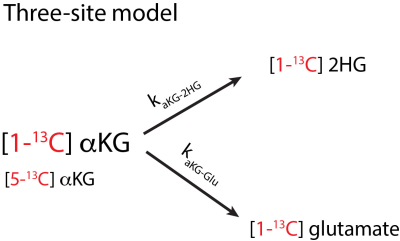 |
A kinetic model to quantify 2-hydroxyglutarate when using hyperpolarized [1-13C]α-ketoglutarate to detect mutant IDH1 in low grade gliomas
Manushka V. Vaidya1, Donghyun Hong1, Sule Sahin1, Georgios Batsios1, Pavithra Viswanath1, Sabrina M. Ronen1, and Peder E.Z. Larson1
1Department of Radiology, University of California San Francisco, San Francisco, CA, United States
We introduce a kinetic modeling framework to detect glutamate and 2-hydroxyglutarate (2HG) production from hyperpolarized [1-13C]alpha-ketoglutarate (C1aKG). Detection of 2HG in vivo is often confounded with [5-13C]alpha-ketoglutarate (C5aKG), a natural abundance peak. We employ the model, based on the solution to differential equations of a three-site model, to separately detect 2HG from C5aKG. To test the model, we used cell lysate data where separate signal peaks of 2HG and C5aKG were experimentally measured. Cases with inputs of 2HG alone, 2HG+C5aKG, and C5aKG alone were evaluated to validate the model.
|
|||
2845.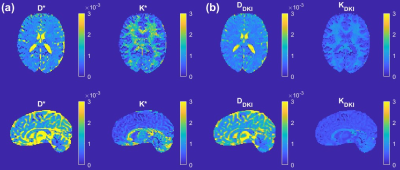 |
A direct link between the DKI model and the sub-diffusion process
Qianqian Yang1 and Viktor Vegh2,3
1Queensland University of Technology, Brisbane, Australia, 2The University of Queensland, Brisbane, Australia, 3Centre for Innovation in Biomedical Imaging Technology, Brisbane, Australia
Diffusion kurtosis imaging (DKI) is an important tool in tissue microstructure studies. The DKI formula for diffusion-weighted MRI signal decay arises through a high order expansion, and the kurtosis has been shown to be sensitive to changes in tissue microstructure. Interestingly, the kurtosis formula describes deviation away from mono-exponential signal decay, like that previously described for anomalous diffusion. Here, we make a link between anomalous sub-diffusion in tissue and diffusion kurtosis. This direct link enables the apparent diffusivity and kurtosis to be computed easily from the sub-diffusion model parameters, leading to superior white-grey matter contrast compared with standard DKI parameters.
|
|||
2846.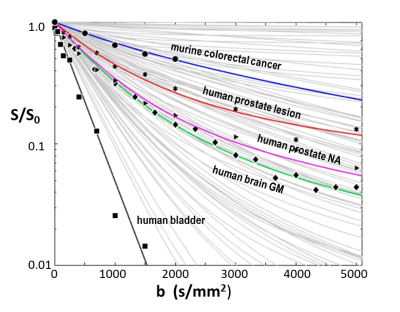 |
Elaborating and Testing Activity MRI [aMRI] Diffusion Modeling
Brendan Moloney1, Xin Li1, Eric M. Baker1, and Charles S. Springer1
1Advanced Imaging Research Center, Oregon Health & Science University, Portland, OR, United States
Activity MRI [aMRI] simulates diffusion-weighted data with three metabolic and cytometric tissue properties: kio (s-1), the mean steady-state cellular water efflux rate constant [measuring cellular metabolic activity], r, the cell density (cells/μL), and V (pL), the average cell volume. We explore the behavior of the aMRI model, and compare its results with pertinent experimental data. The model is well-behaved, and matches experimental data with parameter values in near absolute agreement with independent literature measurements.
|
|||
2847.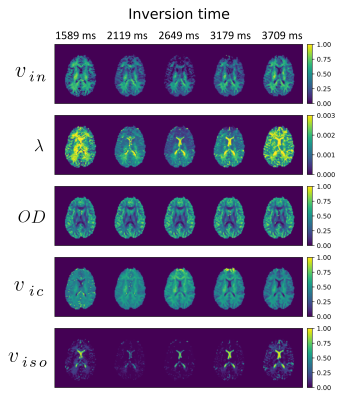 |
The effect of inversion time on a two-compartment SMT and NODDI: an in vivo study
Dominika Ciupek1, Maryam Afzali2, Fabian Bogusz1, Marco Pizzolato3,4, Derek K. Jones2, and Tomasz Pięciak1,5
1AGH University of Science and Technology, Kraków, Poland, 2Cardiff University Brain Research Imaging Centre (CUBRIC), School of Psychology, Cardiff University, Cardiff, United Kingdom, 3Department of Applied Mathematics and Computer Science, Technical University of Denmark, Kongens Lyngby, Denmark, 4Signal Processing Lab (LTS5), École polytechnique fédérale de Lausanne (EPFL), Lausanne, Switzerland, 5LPI, ETSI Telecomunicación, Universidad de Valladolid, Valladolid, Spain
We investigate the effect of the inversion time on the spherical mean technique (SMT) and neurite orientation dispersion and density imaging (NODDI) metrics based on the multi-parametric diffusion MR data. Our findings indicate the orientation dispersion index and mean orientation of Watson distribution remains unchanged for a wide range of TI, while volume fractions and diffusivities show significant changes leading to characteristic "up-and-down" behavior concerning the TI used to fit the model.
|
|||
2848.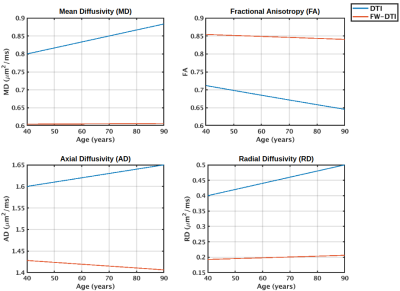 |
Implications of a constant tissue-trace constraint on the two-compartment free water model
Jordan A. Chad1,2, Ofer Pasternak3, and J. Jean Chen1,2
1Department of Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Rotman Research Institute, Baycrest Health Sciences, Toronto, ON, Canada, 3Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States
It has been found that when the single-shell two-compartment free water fit is initialized with a constant trace of the tissue tensor, the final results maintain this constant tissue-trace. It is therefore important to understand the implications of the constant tissue-trace constraint in order to interpret the results of single-shell free water studies. Here we demonstrate that this constraint results in allotting all variation in isotropic diffusivity to the free water compartment while the tissue tensor is unaffected by isotropic variations. It is further shown that the isotropic compartment is more linearly aligned with quadratic variations in diffusivity.
|
|||
2849.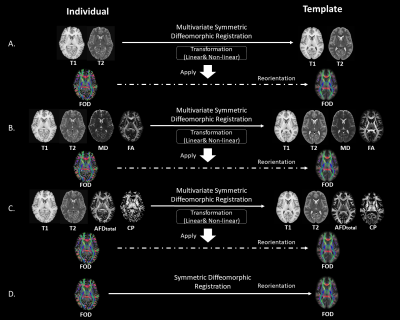 |
Towards an Unbiased Brain Template of Fiber Orientation Distribution Using Multimodal Registration
Jinglei Lv1, Rui Zeng1, and Fernando Calamante1
1School of Biomedical Engineering, The University of Sydney, Sydney, Australia
Building a brain template of fiber orientation distribution (FOD) with Diffusion MRI is crucial for population study and disease research on white matter. The population template and “Fixel” based analysis pipeline is increasingly being used for group-wise statistics. The template generated based solely on symmetric diffeomorphic registration of FOD depicts the group-consistent major fiber bundles; however, spatial specificity is far from optimal in regions near cortical gray matter. In this work, we explore the possibility to leverage the complementary information from T1, T2 and Diffusion MRI and build an unbiased human brain FOD template with multimodal registration method.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.