Digital Posters
Data Processing for Quality & Efficiency
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 15:00 - 16:00 |
3733.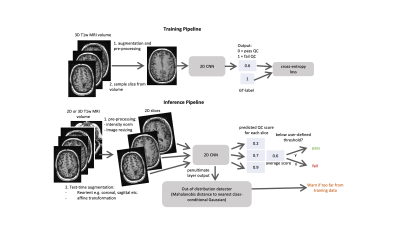 |
Robust and Generalizable Quality Control of Structural MRI images
Ben A Duffy1, Srivathsa Pasumarthi Venkata1, Long Wang1, Sara Dupont1, Lei Xiang1, Greg Zaharchuk1, and Tao Zhang1
1Subtle Medical Inc., Menlo Park, CA, United States
We present an automated deep learning-based quality control system that generalizes to images of different orientations, images with and without contrast as well as those from different acquisition sites. Because the same model was able to classify images with different orientations, test-time augmentation substantially improved performance. Images that were moderately affected by artifacts were able to be identified with 95% accuracy. Furthermore, robustness to different data types (and potentially artifact types) was ensured by using an out-of-distribution detection procedure. This was able to discriminate spine MRI images from T1 brain images with an AUC of 0.98.
|
|||
3734.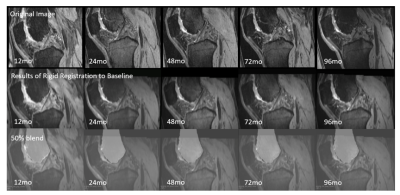 |
Longitudinal Registration of Knee MRI Based on Femoral and Tibial Alignment
Zhixuan Liang1, Yin Guo2, and Chun Yuan3
1Electrical Engineering, Zhejiang University, Hangzhou, China, 2Bioengineering, University of Washington, Seattle, WA, United States, 3Radiology, University of Washington, Seattle, WA, United States
This study aims to develop an automatic and robust algorithm for longitudinal registration of knee Magnetic Resonance Imaging (MRI) across the time span of eight years. We propose a technique that firstly achieves rigid registration based on femoral segmentation, and then makes evaluations based on tibial alignment. Both qualitative and quantitative results show solid performance of the proposed algorithm. This registration algorithm lays a solid foundation for serial analysis of knee images taken over a multi-year time frame.
|
|||
3735.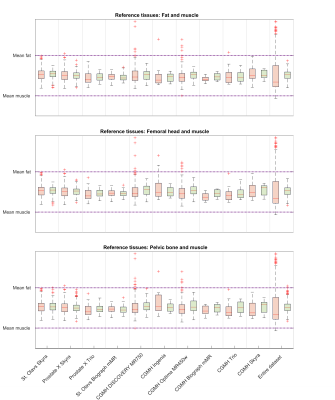 |
Automated reference tissue normalization of prostate T2-weighted MRI on a large, multicenter dataset
Kaia Ingerdatter Sørland1, Mohammed R. S. Sunoqrot1, Pål Erik Goa2,3, Elise Sandsmark3, Sverre Langørgen3, Helena Bertilsson4,5, Gigin Lin6, Tone F. Bathen1,3, and Mattijs Elschot1,3
1Department of Circulation and Medical Imaging, Norwegian University of Science and Technology, Trondheim, Norway, 2Department of Physics, Norwegian University of Science and Technology, Trondheim, Norway, 3Department of Radiology and Nuclear Medicine, St. Olavs University Hospital, Trondheim, Norway, 4Department of Cancer Research and Molecular Medicine, Norwegian University of Science and Technology, Trondheim, Norway, 5Department of Urology, St. Olavs University Hospital, Trondheim, Norway, 6Department of Medical Imaging and Intervention, Chang Gung Memorial Hospital at Linkou, Institute for Radiological Research, Chang Gung University, Taoyuan, Taiwan
Scanner-dependent variations induce non-standard signal intensities (SI) in T2-weighted (T2W) MR images. These variations make computer aided diagnosis of prostate cancer based on T2W images challenging, and SI normalization is necessary. Autoref is a normalization method for axial T2W prostate MRI based on two reference tissues of high and low intensity. The aim of this work was to evaluate Autoref’s performance on a large dataset, and to investigate its performance for various reference tissues. Femoral head and fat were proven to be stable reference tissues, significantly reducing inter-scan variation.
|
|||
3736.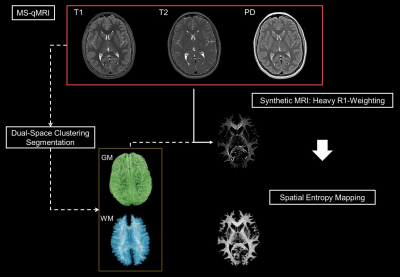 |
qVision for the ELGAN-ECHO Study: An MS-qMRI Processing Pipeline Applied to Large-scale, Multi-site, and Multi-vendor Analyses.
Ryan McNaughton1, Hernan Jara1,2, Chris Pieper2, Laurie Douglass2, Rebecca Fry3, Karl Kuban2, and T. Michael O'Shea3
1Boston University, Boston, MA, United States, 2Boston University Medical Center, Boston, MA, United States, 3University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, MA, United States
Purpose: To describe an integrated, semi-automated image processing pipeline for multispectral qMRI, termed qVision. Methods: Dual-clustering and MS-qMRI python algorithms for the Tri-TSE pulse sequence are automatically calculated and harmonized across a dataset of neuroimaging data from adolescents born extremely preterm. Results: Automated processing is completed in 30 minutes per subject, resulting in high-resolution mappings of T1, T2, PD, and spatial entropy, as well as heavily R1-weighted images of white matter texture via Synthetic-MRI. Conclusion: qVision has been validated on a large-scale, multi-site, and multi-vendor dataset of neuroimaging data, capable of producing a broad spectrum of MS-qMRI outcomes.
|
|||
3737.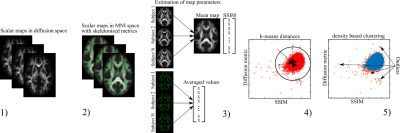 |
YTTRIUM: QC algorithm for the processed diffusion maps in UK Biobank 18608 sample
Ivan I. Maximov1,2, Dennis van der Meer2, Ann-Marie de Lange2, Tobias Kaufmann2, Alexey Shadrin2, Oleksandr Frei2, Thomas Wolfers2, and Lars T Westlye2
1Western Norway University of Applied Sciences, Bergen, Norway, 2NORMENT, University of Oslo, Oslo, Norway
Diffusion MRI is a powerful approach to quantify brain architecture. However, diffusion scalar maps derived from raw data are sensitive to the data quality and processing choices. Many quality control algorithms exist that perform a robust check of raw diffusion data, there is a lack of QCs for inspecting the derived maps from different diffusion approaches. We present a novel QC algorithm for processed scalar maps using mean skeleton values (in the context of tract-based spatial statistics) and structural similarity metric based on the scalar maps. The algorithm builds on clustering of scalar diffusion metrics from 18609 UK Biobank individuals.
|
|||
3738.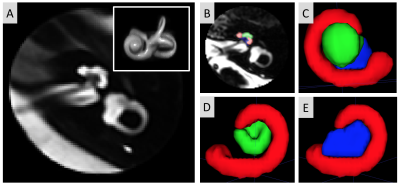 |
Semi-Automated 3D Cochlea Subregional Segmentation on T2-Weighted MRI Scans
William J Matloff1, Daniel J Matloff1, Arthur W Toga1, Taeuk Cheon2, Jangwook Gwak2, Yehree Kim2, Hong Ju Park2, and Hosung Kim1
1Laboratory of Neuro Imaging, USC Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine, University of Southern California, Los Angeles, CA, United States, 2Department of Otorhinolaryngology-Head and Neck Surgery, Asan Medical Center, Seoul, Korea, Republic of
For MRI-based studies of cochlea-related diseases, segmenting the cochlea and modiolus is often essential for quantifying disease-related morphological and intensity changes. Methods for automatic 3D segmentation of these structures on MRI scans, however, are not well established. We aimed to develop a semi-automated, multi-atlas based approach to segmenting the cochlea and modiolus, distinguishing between the cochlear basal and mid-apical turns. We found this approach to have good agreement with manual segmentation, demonstrating its potential in quantitative analyses. Moreover, the definition of the three subregions had good validity, as indicated by the high inter-rater agreement in segmentation.
|
|||
3739.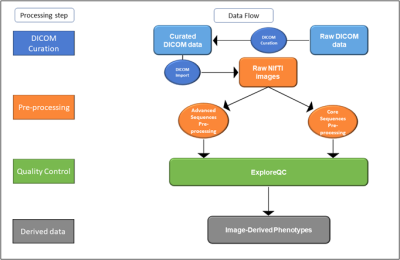 |
Neuroimaging Pre-Processing and Quality Control for The European Prevention of Alzheimer’s Dementia (EPAD) Cohort Study
Luigi Lorenzini1, Silvia Ingala1, Alle Meije Wink1, Joost PA Kuijer1, Viktor Wottschel1, Carole Sudre2,3,4,5, Sven Haller6,7, José Luis Molinuevo8,9,10,11, Juan Domingo Gispert8,10,11,12, David M Cash13, David L Thomas14, Sjoerd B Vos14,15, Ferran Prados Carrasco16,17,18,
Jan Petr19, Robin Wolz20,21, Alessandro Palombit20, Adam J Schwarz22, Gael Chételat23, Pierre Payoux24,25, Carol Di Perri21, Cyril Pernet26, Frisoni Giovanni27,28, Nick C Fox13, Craig Ritchie29, Joanna Wardlaw26,30, Adam Waldman26,31,
Frederik Barkhof1,32, and Henk JMM Mutsaerts1,33
1VUmc Amsterdam, Amsterdam, Netherlands, 2MRC unit for Lifelong Health and Ageing at UCL, London, London, United Kingdom, 3Department of Neurodegenerative Disease, Dementia Research Centre, London, United Kingdom, 4Centre for Medical Image Computing UCL, London, United Kingdom, 5School of Biomedical Engineering & Imaging Sciences, King’s College London, London, United Kingdom, 6CIRD Centre d’Imagerie Rive Droite, Geneva, Switzerland, 7Department of Surgical Sciences, Radiology, Uppsala University, Uppsala, Sweden, 8Barcelonaβeta Brain Research Center (BBRC), Pasqual Maragall Foundation, Barcelona, Spain, 9CIBER Fragilidad y Envejecimiento Saludable (CIBERFES), Madrid, Spain, 10IMIM (Hospital del Mar Medical Research Institute), Barcelona, Spain, 11Universitat Pompeu Fabra, Barcelona, Spain, 12CIBER Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), Madrid, Spain, 13Department of Neurodegenerative Disease, Dementia Research Centre, UCL, London, United Kingdom, 14Neuroradiological Academic Unit, UCL Queen Square Institute of Neurology, London, United Kingdom, 15Centre for Medical Image Computing, University College London, London, United Kingdom, 16Nuclear Magnetic Resonance Research Unit, Queen Square Multiple Sclerosis Centre, University College London Institute of Neurology, London, United Kingdom, 17Department of Medical Physics and Biomedical Engineering, Centre for Medical Image Computing, University College London, London, United Kingdom, 18e-Health Centre, Open University of Catalonia, Barcelona, Spain, 19Helmholtz‐Zentrum Dresden‐Rossendorf, Institute of Radiopharmaceutical Cancer Research, Dresden, Germany, 20IXICO, London, United Kingdom, 21Imperial College London, London, United Kingdom, 22Takeda Pharmaceuticals Ltd., Cambridge, MA, United States, 23Université de Normandie, Unicaen, Inserm, U1237, PhIND "Physiopathology and Imaging of Neurological Disorders", institut Blood-and-Brain @ Caen-Normandie, Cyceron, Caen, France, 24Department of Nuclear Medicine, Toulouse CHU, Purpan University Hospital, Toulouse, France, 25Toulouse NeuroImaging Center, University of Toulouse, INSERM, UPS, Toulouse, France, 26Centre for Clinical Brain Sciences, The University of Edinburgh, Edinburgh, Scotland, 27Laboratory Alzheimer’s Neuroimaging & Epidemiology, IRCCS Istituto Centro San Giovanni di Dio Fatebenefratelli, Brescia, Italy, 28University Hospitals and University of Geneva, Geneva, Switzerland, 29Centre for Dementia Prevention, The University of Edinburgh, Edinburgh, Scotland, 30UK Dementia Research Institute at Edinburgh, University of Edinburg, Edinburgh, Scotland, 31Department of Medicine, Imperial College London, London, United Kingdom, 32Institute of Neurology and Healthcare Engineering, University College London, London, United Kingdom, 33Ghent Institute for Functional and Metabolic Imaging (GIfMI), Ghent University, Ghent, Belgium
The neuroimaging community strives to obtain large data cohorts, usually through association within consortia spanning different sites and countries. This results in increased variability of acquisition parameters and scan quality, which can affect image processing and statistical analyses. We propose a semi-automatic data management pipeline to process raw data, assess quality and compute image-derived phenotypes from multi-modal MRI scans, as developed for the multi-centre European Prevention of Alzheimer Dementia longitudinal cohort study (EPAD LCS).
|
|||
3740.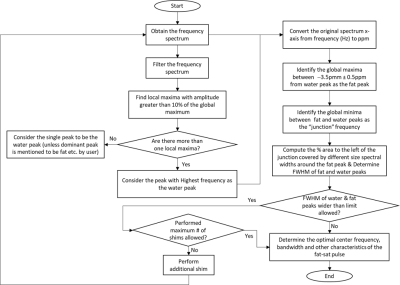 |
Algorithm for Automated Identification of Spectral Characteristics
Venkata Veerendranadh Chebrolu1, Michael Wullenweber2, Andreas Schaefer2, Johann Sukkau2, and Peter Kollasch1
1Siemens Medical Solutions USA, Inc., Rochester, MN, United States, 2Siemens Healthcare GmbH, Erlangen, Germany
Automated identification of proton spectral characteristics has potential utility in accurate spectral fat saturation, improving dynamic shim routines, and optimizing bandwidth of radiofrequency pulses used in multi-slice or multi-band excitation. In this work, we present an algorithm for automated identification of fat and water proton spectral characteristics and evaluate its performance in 30 proton spectra from breast (number of subjects: n=20), ankle (n=11), and knee (n=9) anatomical regions.
|
|||
3741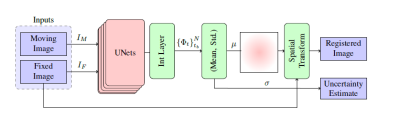 |
Assessment of Uncertainty in Brain MRI Deformable Registration Video Permission Withheld
Samah Khawaled1 and Moti Freiman2
1Applied Mathematics, Technion, Haifa, Israel, 2Biomedical Engineering, Technion, Haifa, Israel
Unsupervised deep neural networks (DNN) are successfully employed to predict deformation-fields in neuroimaging studies. Bayesian DNN models enable safer utilization of DNN methods in neuroimaging studies, improve generalization and enable assessment of uncertainty in the predictions. We propose a non-parametric Bayesian approach to estimate the uncertainty in DNN-based algorithms for brain MRI deformable registration. We demonstrated the added-value of our Bayesian registration framework on the brain MRI (LPBA40) dataset compared to state-of-the-art VoxelMorph DNN. Further, we quantified the uncertainty of the registration and assessed its correlation with the out-of-distribution data.
|
|||
3742.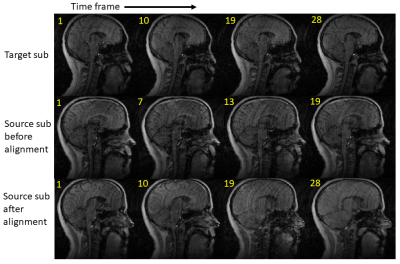 |
Temporal Frame Alignment for Speech Atlas Construction Using High Speed Dynamic MRI
Fangxu Xing1, Riwei Jin2, Imani Gilbert3, Xiaofeng Liu1, Georges El Fakhri1, Jamie Perry3, Bradley Sutton2, and Jonghye Woo1
1Department of Radiology, Massachusetts General Hospital, Boston, MA, United States, 2Department of Bioengineering, University of Illinois at Urbana-Champaign, Champaign, IL, United States, 3Department of Communication Sciences and Disorders, East Carolina University, Greenville, NC, United States
Visual assessment of articulatory motion of the vocal tract in speech has been drastically advanced by recent developments in high-speed real-time MRI. However, the variation of speaking rate among different subjects prevents quantitative analysis of data from a larger study group. We present a pipeline of methods that aligns audio and image data in the time domain and produces temporally matched image volumes for various subjects. Comparison of the cross-correlation score before and after time alignment showed an increased similarity between source and target image sequences, enabling production of preprocessed multi-subject data for the subsequent statistical atlas construction studies.
|
|||
3743.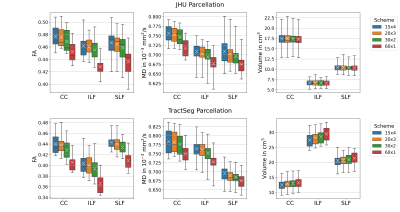 |
Reproducibility of White Matter Parcellation on Multi-Acquisition Diffusion Weighted Imaging
Stefan Winzeck1,2, Ben Glocker1, Virginia F. J. Newcombe2, David K. Menon2, and Marta M. Correia3
1BioMedIA, Department of Computing, Imperial College London, London, United Kingdom, 2Division of Anaesthesia, Department of Medicine, University of Cambridge, Cambridge, United Kingdom, 3MRC Cognition and Brain Sciences Unit, University of Cambridge, Cambridge, United Kingdom
The comparison of TractSeg and JHU atlas-based white matter parcellation on DWI showed the impact of different acquisition schemes on both region volumes and variation in diffusion metrics. TractSeg defined fibre bundles more accurately, but volumes varied for different DWI parameters. The atlas-based segmentation of fibre tracts was more robust to acquisition differences, but showed higher variation in diffusion metrics suggesting a less precise differentiation of white and grey matter.
|
|||
3744.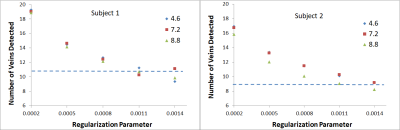 |
Performance evaluation of a Compressed Sensing SWI technique on a clinical 7T MRI system
Emily Koons1, Eric G Stinson2, Patrick Liebig3, Peter Speier3, Krystal Kirby1, Kirk M Welker1, and Andrew J Fagan1
1Radiology, Mayo Clinic, Rochester, MN, United States, 2Siemens Medical Solutions USA, Inc., Rochester, MN, United States, 3Siemens Healthcare GmbH, Erlangen, Germany
A compressed sensing technique was adapted for use in acquiring 3D susceptibility-weighted image data on a clinical 7T MRI system. The image quality was highly dependent on the choice of regularization parameter used in the reconstruction for all accelerated acquisitions, with some residual artifacts manifest in images thought due to the acquisition of anisotropic voxels. Nevertheless, CNR and vein detectability was significantly enhanced in images acquired using the CS-SWI technique compared to the clinical SWI protocol, for CS acceleration factors up to 7.2 (corresponding to a 48% scan time reduction compared to the clinical protocol).
|
|||
3745.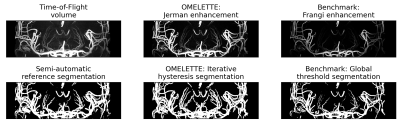 |
Openly available sMall vEsseL sEgmenTaTion pipelinE (OMELETTE)
Hendrik Mattern1
1Biomedical Magnetic Resonance, Otto-von-Guericke-University Magdeburg, Magdeburg, Germany
An easy-to-use and Openly available sMall vEsseL sEgmenTa-Tion pipelinE (OMELETTE) was developed and compared to a Frangi-based benchmark segmentation. Segmentations performance was evaluated for 20 high resolution Time-of-Flight angiograms. Qualitative and quantitative comparison showed OMELETTE's superior segmentation performance.
|
|||
3746.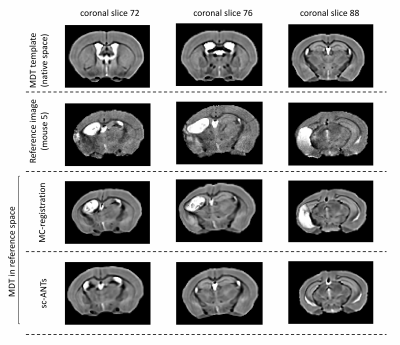 |
A Multicomponent Image Registration Technique for Largely Deformed Ventricles in Mouse Brain After Stroke
Warda T. Syeda1, Vanessa Brait2, Alex Oman2, Charlotte Ermine 2, Jess Nithianantharajah 2, Lachlan Thompson2, Leigh A. Johnston3,4, David K. Wright5, and Amy Brodtmann2
1Melbourne Neuropsychiatry Centre, The University of Melbourne, Parkville, Australia, 2The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Parkville, Australia, 3Department of Biomedical Engineering, The University of Melbourne, Melbourne, Australia, 4Melbourne Brain Centre Imaging Unit, The University of Melbourne, Parkville, Australia, 5Department of Neuroscience, Central Clinical School, Monash University, Melbourne, Australia
A multicomponent registration technique has been proposed to perform image normalization in the presence of largely deformed ventricles in the mouse brain. By employing multicomponent similarity metrics, the proposed technique combines information from multiple filtered and contrast-enhanced copies of the original images in the optimization process. We apply the proposed method to a mouse brain dataset with enlarged ventricles after focal ischaemic stroke, and compare the performance with single-component registration.
|
|||
3747.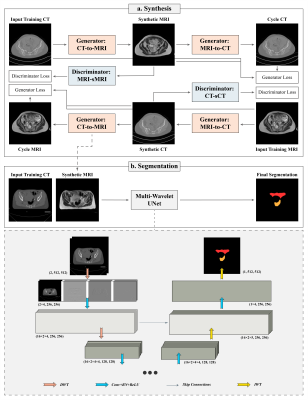 |
Synthetic MRI-assisted Multi-Wavelet Segmentation Framework for Organs-at-Risk Delineation on CT for Radiotherapy Planning
Reza Kalantar1, Susan Lalondrelle2, Jessica M Winfield1,3, Christina Messiou1,3, Dow-Mu Koh1,3, and Matthew D Blackledge1
1Division of Radiotherapy and Imaging, The Institute of Cancer Research, London, United Kingdom, 2Gynaecological Unit, The Royal Marsden Hospital, London, United Kingdom, 3Department of Radiology, The Royal Marsden Hospital, London, United Kingdom
Radiotherapy (RT) is a cornerstone treatment for cervical cancer. Accurate delineation of organs-at-risk (OARs) is critical to prevent radiation toxicity to healthy organs surrounding the tumour. However, OARs delineation is currently performed manually by clinicians which is labour- and time-intensive. Deep-learning-based algorithms have shown potential in automating this task. This study developed and evaluated a novel framework to incorporate the superior soft-tissue contrast of MRI as well as multi-wavelet image decompositions for improved OARs segmentation on CT images. The proposed framework appears to be a promising addition to the cervical cancer treatment workflow.
|
|||
3748.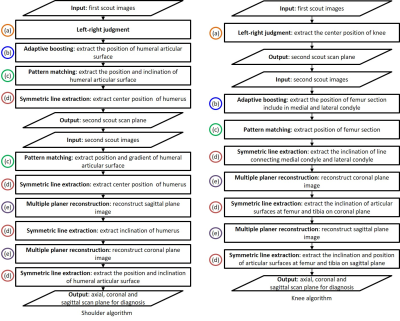 |
Automated scan plane planning for multiple examination parts by modular algorithm developing method
Suguru Yokosawa1, Toru Shirai1, Hisako Nagao1, and Hisaaki Ochi1
1Healthcare Business Unit, Hitachi, Ltd, Tokyo, Japan
Generally, automated scan plane planning methods require the recognition of different landmarks depending on the examination parts. Therefore, algorithms must be tailored to each examination part. In this study, we have proposed a modular algorithm developing method for automatic scan plane planning. In this method, an algorithm is composed of several common processes, and by simply changing the combination of those processes, it can be tailored to different examination parts.
|
|||
3749.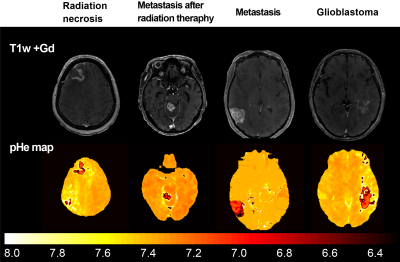 |
Brain Tumor Characterization and Assessment using Automatic Detection of Extracellular pH Change
Yuki Matsumoto1, Masafumi Harada1, Yuki Kanazawa1, Nagomi Fukuda2, Syun Kitano2, Yo Taniguchi3, Masaharu Ono3, and Yoshitaka Bito3
1Graduate School of Biomedical Sciences, Tokushima University, Tokushima-city, Japan, 2School of Health Sciences, Tokushima University, Tokushima-city, Japan, 3Healthcare Business Unit, Hitachi, Ltd., Tokyo, Japan
In this study, we attempted to characterize brain tumors by combining quantitative parameter mapping and deep-learning-based semantic segmentation. T1, concentration of contrast media (CM), and pHe maps were calculated after the image dataset was obtained. The contrast-enhanced area was then automatically detected using a deep-learning-based semantic segmentation algorithm. The segmented mask was set as the region of interest on these calculated maps. The statistical significance of differences in brain tumors was evaluated to determine whether changes in the mean T1, CM, and pHe were malignancy-dependent.
|
|||
3750.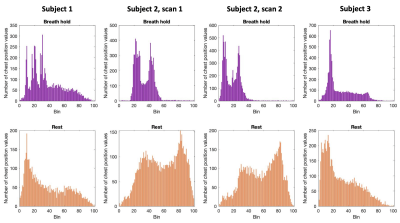 |
Adding an absolute chest position regressor to RETROICOR for spinal cord fMRI during atypical breathing patterns
Neha A Reddy1,2, Andrew D Vigotsky1,3, Rachael C Stickland2, and Molly G Bright1,2
1Department of Biomedical Engineering, Northwestern University, Evanston, IL, United States, 2Department of Physical Therapy and Human Movement Sciences, Feinberg School of Medicine, Northwestern University, Chicago, IL, United States, 3Department of Statistics, Northwestern University, Evanston, IL, United States
RETROICOR (Retrospective Image Correction) is a commonly used technique for physiological noise correction, but it may not be optimal in its standard implementation for use in studies of the human spinal cord involving atypical breathing patterns. We test the inclusion of an absolute chest position regressor derived from respiratory belt data, in addition to standard RETROICOR regressors, in modeling fMRI scans of the cervical spinal cord with and without breath-hold challenges. The chest position regressor may be a simple approach towards explaining additional noise variance in spinal cord fMRI data during breath-hold tasks.
|
|||
3751.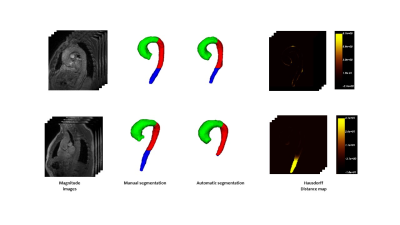 |
Automatic 3D PC-MRI atlas-based segmentation of the aorta
Diana M. Marin-Castrillon1, Arnaud Boucher1, Siyu Lin1, Chloe Bernard2, Marie-Catherine Morgant1,2, Alexandre Cochet1,3, Alain Lalande 1,3, Benoit Presles 1, and Olivier Bouchot 1,2
1ImViA Laboratory, University of Burgundy, Dijon, France, 2Department of Cardio-Vascular and Thoracic Surgery, University Hospital of Dijon, Dijon, France, 3Department of Medical Imaging, University Hospital of Dijon, Dijon, France
Analysis of aorta hemodynamics is useful in the evaluation of aortic diseases. 4D PC-MRI provides information of flow velocity in the aorta and automatic segmentation is one of the biggest challenges. We propose a fully 3D automatic segmentation of the aorta in systole using a multi-atlas approach. Evaluation on 16 patients provided an average performance of 29.55±24.33 mm and 0.859 ±0.024 for Hausdorff distance and Dice score respectively. With the proposed method, the automatic segmentation of the thoracic aorta that can be obtained from 4D PC-MRI is close enough to the manual one to be used in future studies.
|
|||
3752.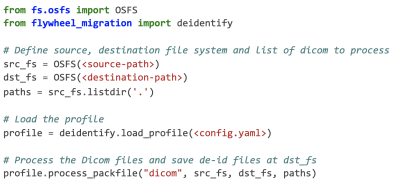 |
A flexible open-source Python package for de-identification of medical images and related data
Nicolas Pannetier1, Kaleb Fischer1, Justin Elhert1, Ambrus Simon1, Gunnar Schaefer1, and Michael Perry1
1Flywheel Exchange, Inc, Minneapolis, MN, United States
De-identification of medical data is complex and is a barrier for aggregating heterogeneous data repositories. We have developed a flexible open-source Python package for de-identification of medical images and related data. It is fully featured, supports DICOM and 8 other file types and 9 different field transformations. This package is used in production at Flywheel, Inc. We believe this package can contribute to enforce best standards in the protection of patient identity and privacy while relieving researchers from the burdensome task of developing their own custom tooling.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.