Digital Posters
Modelling, Reconstruction & Processing in Low-Field MRI & PET-MRI
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 17:00 - 18:00 |
 |
2182.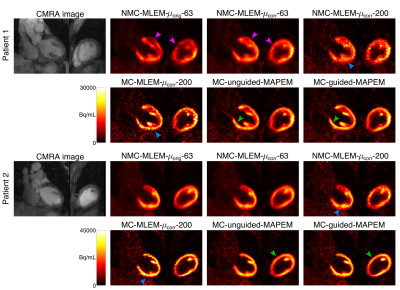 |
MR-based motion correction and anatomical guidance for improved PET image reconstruction in cardiac PET-MR imaging
Camila Munoz1, Sam Ellis1, Stephan G Nekolla2, Karl P Kunze1,3, Teresa Vitadello4, Radhouene Neji1,3, René M. Botnar1, Julia A. Schnabel1, Andrew J. Reader1, and Claudia Prieto1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Nuklearmedizinische Klinik und Poliklinik, Technische Universitat Munchen, Munich, Germany, 3MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom, 4Department of Internal Medicine I, University hospital rechts der Isar, Technical University of Munich, Munich, Germany
Simultaneous PET-MR has shown promise for addressing several of the technical challenges that may degrade image quality in PET imaging, such as high noise levels, attenuation artefacts, and motion artefacts. While state-of-the-art PET image reconstruction techniques have addressed these issues separately, their combined effect has not been demonstrated. Here we introduce a single framework that integrates MR-based motion correction and anatomical guidance for improved simultaneous diagnostic cardiac PET-MR imaging. We evaluated the proposed framework on cardiac [18F]FDG PET-MR datasets and results show that, compared to conventional reconstruction algorithms, our framework results in sharper images, with increased contrast and reduced noise.
|
||
2183.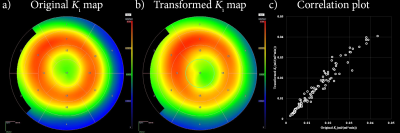 |
Impact of motion on simultaneously acquired PET/MRI of myocardial infarcted heart.
Heeseung Lim1, Benjamin Wilk 1,2, Jane Sykes 1, John Butler 1, Gerald Moran3, Jonathan Thiessen1,2, Gerald Wisenberg1,4, and Frank S Prato1,2
1Lawson Health Research Institute, London, ON, Canada, 2Medical Biophysics, Western University, London, ON, Canada, 3Siemens Healthcare Limited, Oakville, ON, Canada, 4MyHealth Centre, Arva, ON, Canada
This study investigates the impact of motion in myocardial PET/MRI data by registering pre-mortem images to post-mortem images from a simultaneously acquired PET/MRI scan. After a registration in MRI, PET data is transformed and analyzed using the Patlak model. There are significant differences and correlation between pre- and post-mortem PET images. There are discrepancies in different segments of heart, but none were found to be significantly different. These results suggest that the level of inflammation in the heart might be misrepresented. Further comparison with motion corrected images will provide more concrete discrepancy due to motion for myocardial PET/MRI imaging.
|
|||
2184.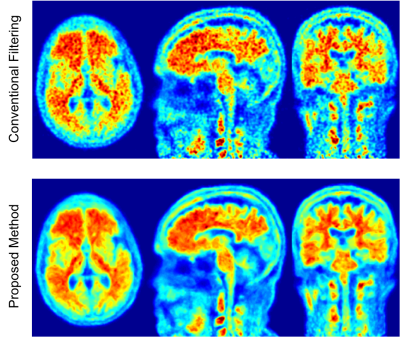 |
High resolution PET image denoising using anatomical priors by K-nearest neighborhood method in the feature space
Mehdi Khalighi1, Timothy Deller2, Kevin Chen1, Tyler Toueg3, Dawn Holley1, Kim Halbert1, Floris Jansen2, Elizabeth Mormino3, Michael Zeineh1, Farshad Moradi1, Greg Zaharchuk1, and Andrei Iagaru1
1Radiology, Stanford University, Stanford, CA, United States, 2Engineering Dept., GE Healthcare, Waukesha, WI, United States, 3Neurology, Stanford University, Stanford, CA, United States
After PET images are reconstructed by OSEM, a spatial filter which exploits the correlation between neighboring voxels, is applied to remove noise. A new filtering method is proposed that also exploits the correlation between voxels from the same tissue. Anatomical priors are processed for bias correction and registered to PET images. For each voxel, similar voxels within the PET volume are identified using KNN method in the feature-space built by anatomical priors. These similar voxels and also the neighboring voxels are then used to remove the high frequency noise on PET images using a Gaussian and a weighted averaging filter.
|
|||
2185.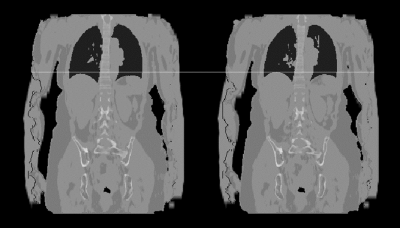 |
Free-Breathing MR-based Attenuation Correction for Whole-Body PET/MR Exams
Patrick Korf1, Wolfgang Thaiss2, Ambros J. Beer2, Meinrad Beer3, Dominik Nickel1, and Thomas Vahle1
1Siemens Healthcare GmbH, Erlangen, Germany, 2Department of Nuclear Medicine, University Hospital Ulm, Ulm, Germany, 3Department of Diagnostic and Interventional Radiology, University Hospital Ulm, Ulm, Germany
In whole-body PET/MR exams, MR-based attenuation correction is usually performed with a Dixon protocol of an MR VIBE sequence acquired in breath-hold followed by a segmentation into different tissue classes. As an extension we present a free-breathing approach for attenuation correction that can be used for patients that have problems or are even unable to perform the required breath-holds. The presented approach relies on a self-gated, compressed sensing accelerated gradient-echo sequence with Cartesian k-space sampling. We demonstrate the generation of free-breathing attenuation maps in 2 human volunteers and 10 patients.
|
|||
2186.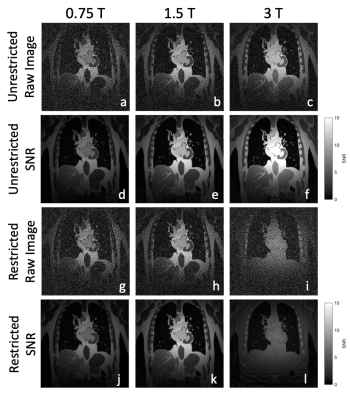 |
First-Principle Image SNR Synthesis Depending on Field Strength
Charles McGrath1, Mohammed M Albannay1, Alexander Jaffray1, Christian Guenthner1, and Sebastian Kozerke1
1Institute for Biomedical Engineering, University and ETH Zurich, Zurich, Switzerland
The signal-to-noise ratio (SNR) is a key metric of imaging performance, however many formulations do not account for relaxation properties and sequence timing effects, which play a decisive role when studying the effect of static field strength $$$B_0$$$ on SNR. We have developed first-principle simulations that incorporate these effects in order to estimate SNR scaling relations, and show that SNR can deviate significantly from previous scaling laws, specifically for lower field strengths and when sequence restrictions apply.
|
|||
2187.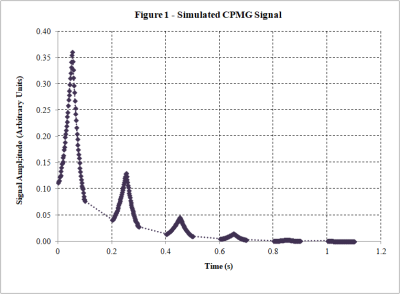 |
Development of a Numerical Bloch Solver for Low-Field Pulse Sequence Modeling
John Adams1,2, William Handler1,2, and Blaine Chronik1,2
1Department of Physics and Astronomy, Western University, London, ON, Canada, 2xMR Labs, London, ON, Canada
Renewed interest in clinical low-field MR systems has opened up a new design space for MR pulse sequence design. To explore these opportunities, and to better inform hardware design, we are developing a flexible simulation tool based on a numerical simulation of the Bloch equations. This tool will both be able to model pulse sequences under the influence of realistic applied fields, and account for changes in relaxation time due to changes in field strength. This abstract presents our work to date in developing this tool.
|
|||
2188.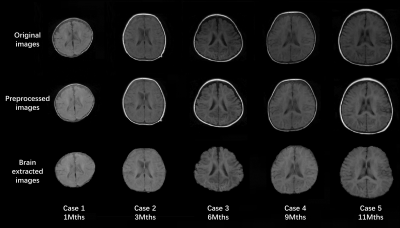 |
Automatic Quantitative Analysis of Low-field Infant Brain MR Images
Bo Peng1,2,3, Baohua Hu1,2,3, Mao Sheng4, Yuqi Liu4, Zhongchang Miao5, Zijun Dong6, Jian Bao7, SiSeung Kim7, Bing Keong Li7, and Yakang Dai1,2,3
1Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou, China, 2Suzhou Key Laboratory of Medical and Health Information Technology, Suzhou, China, 3Jinan Guoke Medical Engineering Technology Development co., Ltd., Jinan, China, 4Department of Radiology, Children’s Hospital of Soochow University, Suzhou, China, 5Department of Radiology, The First People’s Hospital of Lianyungang, Jiangsu Province, China, 6Department of Medical Imaging, Lianyungang Women and Children Hospital and Health Institute, Jiangsu Province, China, 7Jiangsu LiCi Medical Device Co., Ltd., Lianyungang, China
Low-field MRI is foreseeable as a safer system for infants. However, low-field MR images have lower SNR and spatial resolution as compared to high-field images, thus processing of low-field infant brain MR image is challenging. In this study, an automated image processing method that can accurately perform brain extraction, tissue segmentation, and brain labeling on low-field infant brain MR images is developed. It is also capable to automatically construct the inner, middle, and outer surfaces of the cerebral cortex and provides automatic quantitative analysis of selected region of interest, which can be a helpful tool for researchers in neuroimaging studies.
|
|||
2189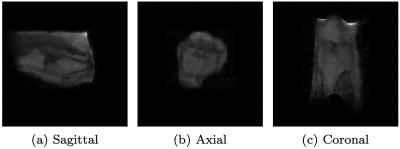 |
Low-field MR imaging using multiplicative regularization Video Permission Withheld
Merel de Leeuw den Bouter1, Martin van Gijzen1, and Rob Remis2
1Delft Institute of Applied Mathematics, Delft University of Technology, Delft, Netherlands, 2Circuits and Systems, Delft University of Technology, Delft, Netherlands We present an image reconstruction approach that incorporates regularization by multiplying the data-fidelity term by a total variation functional. Usually, regularization is carried out in an additive manner, with a regularization parameter balancing out the two terms. Such a parameter often needs to be tuned for each dataset through extensive numerical experimentation. Our approach does not require such a parameter. We applied the method to in-vivo data acquired using a low-field MR scanner. Our results show that the method successfully denoises low-field MR images. |
|||
2190.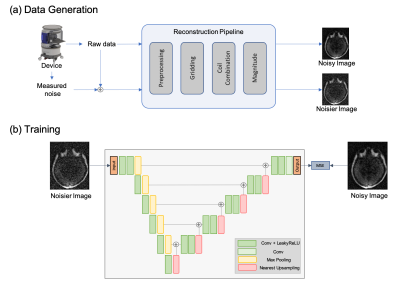 |
Unsupervised Denoising for Low-field Diffusion MRI
Jo Schlemper1, Neel Dey2, Seyed Sadegh Mohseni Salehi1, Carole Lazarus1, Rafael O'Halloran1, Prantik Kundu1,3, and Michal Sofka1
1Hyperfine Research Inc., Guilford, CT, United States, 2New York University, New York, NY, United States, 3Icahn School of Medicine at Mount Sinai, New York, NY, United States
An unsupervised deep learning framework is proposed for denoising low-field 64 mT diffusion-weighted MRI images (DWI). The denoised DWI (b-value = 890 s/mm2) and apparent diffusion coefficient (ADC) maps were evaluated in a user study by four expert graders in terms of sharpness, noise reduction, and overall utility. Our framework was found to enable low-field DWI restoration with strong noise while maintaining relevant image features. 62.50% and 64.28% of processed images were rated clearly/far better overall for DWI and ADC, respectively, with only 0.05% of processed DWI and 0% of processed ADC rated clearly/far worse.
|
|||
2191.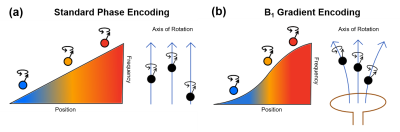 |
Correction of Image Distortions Arising from RF Encoding with Nonlinear Fields
Paul Wang1, Michael Mullen2, Lance DelaBarre2, and Michael Garwood2
1Center for Magnetic Resonance Research and Department of Biomedical Engineering, University of Minnesota, Minneapolis, MN, United States, 2Center for Magnetic Resonance Research and Department of Radiology, University of Minnesota, Minneapolis, MN, United States
In this work, we investigate whether an established method to correct distortions in conventional MRI can be repurposed to correct distortions arising from the nonlinearity of B1 gradients when performing RF-encoded MRI at low field, where SAR constraints are reduced. Although several methods are capable of correcting image distortions arising from nonlinear B0 gradients and/or large B0 inhomogeneity, here we chose to adapt the method of Weis et al. Through theory and simulations, we demonstrate the ability to correct image distortions arising from nonlinear B1 gradients in RF-encoded MRI.
|
|||
2192.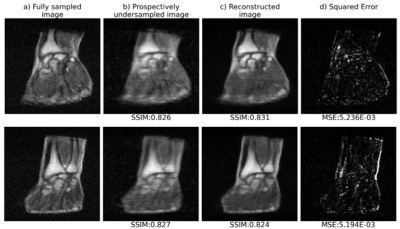 |
Deep learning for fast 3D low field MRI
Reina Ayde1, Tobias Senft1, Najat Salameh1, and Mathieu Sarracanie1
1Center for Adaptable MRI Technology (AMT Center), Department of Biomedical Engineering, University of Basel, Allschwil, Switzerland
Low magnetic field (LF) MRI is currently gaining momentum as a complementary, more flexible and cost-effective approach to MRI diagnosis. However, the impaired Signal-to-Noise Ratio, leading in turn to prolonged acquisition times, challenges its relevance at the clinical level. Recently, reconstructing an alias-free image using deep learning techniques has shown promising results. In this study, we leverage deep learning reconstruction to demonstrate the feasibility of highly undersampled (20% sampling) 3D LF MRI at 0.1 T. The model performance has been evaluated on both retrospective and acquired, prospective 3D LF data.
|
|||
 |
2193.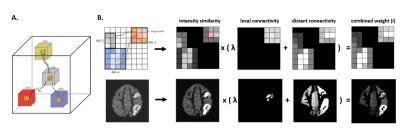 |
CONN-NLM: a novel CONNectome-based Non-Local Means filter for PET-MRI denoising
Zhuopin Sun1, Steven Meikle2,3, and Fernando Calamante1,3,4
1School of Biomedical Engineering, The University of Sydney, Sydney, Australia, 2Faculty of Medicine and Health, The University of Sydney, Sydney, Australia, 3Brain and Mind Centre, The University of Sydney, Sydney, Australia, 4Sydney Imaging, The University of Sydney, Sydney, Australia
Recent advances in hybrid PET-MRI systems enable simultaneous acquisition of PET and MR data. PET is used to visualize and measure biochemically-specific metabolic processes, but has limited spatial resolution and signal-to-noise ratio. Combining diffusion MRI (dMRI) and PET data, which provide highly complementary information (e.g. structural connectivity and molecular information), has rarely been exploited previously in image postprocessing. The proposed CONNectome-based Non-Local Means (CONN-NLM) exploits synergies between dMRI-derived structural connectivity and PET intensity information to denoise PET data. This method is based on the rationale that structurally-connected voxels and voxels with similar intensity should be highly weighted when smoothing noise.
|
||
2194.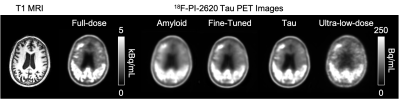 |
Generalizing Ultra-low-dose PET/MRI Networks Across Radiotracers: From Amyloid to Tau
Kevin T. Chen1, Olalekan Adeyeri2, Tyler N Toueg3, Elizabeth Mormino3, Mehdi Khalighi1, and Greg Zaharchuk1
1Radiology, Stanford University, Stanford, CA, United States, 2Salem State University, Salem, MA, United States, 3Neurology and Neurological Sciences, Stanford University, Stanford, CA, United States
We have previously trained a deep learning network with simultaneous PET/MRI inputs to generate diagnostic quality images from ultra-low-dose amyloid PET acquisitions. With data bias being a known issue in deep learning-based applications, we aim to investigate whether this network could generalize to ultra-low-dose tau PET image enhancement. Results of this study show that data bias across radiotracers needs to be accounted for before applying an ultra-low-dose network trained on one tracer to another.
|
|||
2195.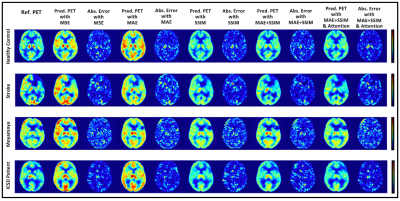 |
Ablation Studies in 3D Encoder-Decoder Networks for Brain MRI-to-PET Cerebral Blood Flow Transformation
Ramy Hussein1, Moss Zhao1, Jia Guo2, Kevin Chen1, David Shin3, Michael Moseley1, and Greg Zaharchuk1
1Radiology, Stanford University, Stanford, CA, United States, 2Bioengineering, University of California, Riverside, Riverside, CA, United States, 3Neuro MR, GE Healthcare, Menlo Park, CA, United States
In this study, we demonstrate that an optimized 3D encoder-decoder structured convolutional neural network with attention gates can effectively integrate brain structural MRI and ASL perfusion images to produce high-quality synthetic PET CBF maps without using radiotracers. We performed experiments to evaluate different loss functions and the role of the attention mechanism. Our results showed that attention-based 3D encoder-decoder network with custom loss function produces the superior PET CBF prediction results, achieving SSIM of 0.94, MSE of 0.00025, and PSNR of 38dB.
|
|||
2196.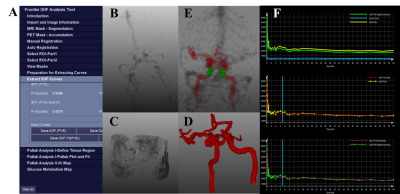 |
Development and Evaluation of a software for Parametric Patlak mapping using PET/MRI input function (CALIPER).
Praveen Dassanayake1,2, Lumeng Cui3, Elizabeth Finger2,4, Andrea Soddu5,6, Bjoern Jakoby7, Keith St. Lawrence1,2, Gerald Moran8, and Udunna Anazodo1,2
1Department of Medical Biophysics, University of Western Ontario, London, ON, Canada, 2Lawson Health Research Institute, London, ON, Canada, 3Division of Biomedical Engineering, University of Saskatchewan, Saskatoon, SK, Canada, 4Department of Clinical Neurological Sciences, University of Western Ontario, London, ON, Canada, 5Brain and Mind Institute, University of Western Ontario, London, ON, Canada, 6Department of Physics and Astronomy, University of Western Ontario, London, ON, Canada, 7Department of Physics, University of Surrey, Guildford, United Kingdom, 8Siemens Healthineers, Oakville, ON, Canada
Absolute quantification of tracer uptake in positron emission tomography (PET) requires the knowledge of an arterial input function (AIF) which involves invasive arterial blood sampling. Alternatively, input functions can be extracted by identifying feeding arteries from PET images. In this study we validated a software that uses magnetic resonance images to identify the feeding arteries in PET images to generate image derived input function (IDIF) for absolute quantification of PET. The ratio of area under curve between IDIFs and AIFs revealed that this tool can generate accurate IDIFs for non-invasive PET quantification.
|
|||
2197.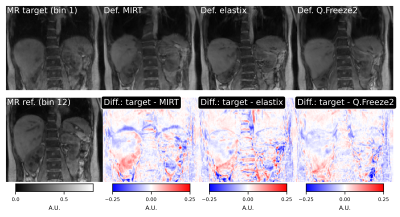 |
Comparison of deformable registration techniques for real-time MR-based motion correction in PET/MR
Thibault Marin1, Yanis Djebra1,2, Paul Han1, Vanessa Landes3, Yue Zhuo1, Kuan-Hao Su4, Georges El Fakhri1, and Chao Ma1
1Massachusetts General Hospital, Harvard Medical School, Boston, MA, United States, 2LTCI, Telecom Paris, Institut Polytechnique de Paris, Paris, France, 3GE Healthcare, Boston, MA, United States, 4GE Healthcare, Waukesha, WI, United States
Motion during acquisition of PET/MR data can severely degrade image quality of PET/MR studies. We have previously reported an MR-based motion correction technique capable of correcting for irregular motion patterns such as bulk motion and irregular respiratory motion. The method is based on a subspace MR model enabling reconstruction of real-time volumetric MR images (9 frames per second). In this work, we present improvements to the motion estimation method used to obtain motion fields from real-time MR images. We compare the performance of three packages for irregular motion patterns.
|
|||
2198.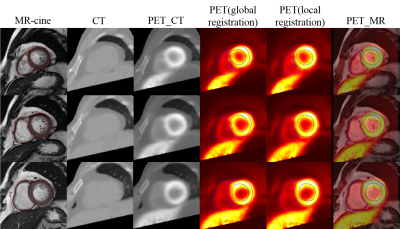 |
A registration approach for cardiac PET/CT and MR images
Xiaomeng Wu1, Shuai Liu1, Li Huo2, Xihai Zhao3, and Fei Shang1
1Department of Biomedical Engineering, School of Life Science, Beijing institute of technology, beijing, China, 2Department of Nuclear Medicine, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China, 3Department of Biomedical Engineering, Center for Biomedical Imaging Research, Tsinghua University School of Medicine, Beijing, China
Multi-modality images can provide comprehensive information for clinical diagnosis. In this study, a registration method for cardiac PET and MR images was proposed by combining global registration and local registration. During global registration, axial PET and CT images were transformed to short axis with MR-survey as link. Then local registration was performed between PET/CT and MR-cine images, it was found that using the fusion of PET and CT images performed better compared with that only using a single modality image.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.