Digital Posters
Multicomponent Models of Diffusion, Perfusion & Relaxation
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 4 | 19:00 - 20:00 |
3421.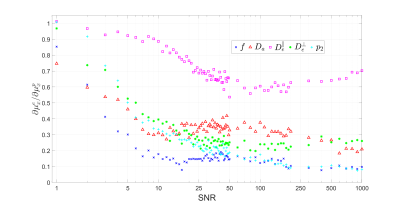 |
Effect of the training set on supervised-learning parameter estimation: Application to the Standard Model of diffusion in white matter
Ying Liao1, Santiago Coelho1, Jelle Veraart1, Els Fieremans1, and Dmitry S. Novikov1
1Radiology, NYU School of Medicine, New York, NY, United States
Maximum likelihood estimation is challenging in multicompartmental models due to the degeneracy of the optimization landscape. As a result, machine learning (ML) methods are often applied for parameter estimation, interpolating the mapping of measurements to model parameters. Such mapping can essentially depend on the training set (prior), decreasing the sensitivity to the measurements, and yielding artificially “clean” maps. Here we quantify the effect of the training set on the Standard Model of diffusion in white matter as function of signal-to-noise ratio, in simulations and in vivo.
|
|||
3422.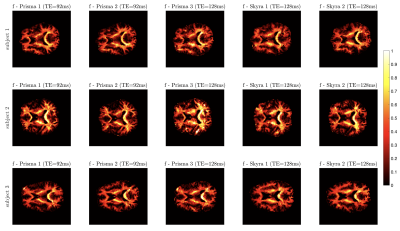 |
Feasibility of white matter Standard Model parameter estimation in clinical settings
Santiago Coelho1, Steven Baete1, Gregory Lemberskiy1, Benjamin Ades-aron1, Jelle Veraart1, Dmitry S. Novikov1, and Els Fieremans1
1Radiology, NYU School of Medicine, New York, NY, United States
Robust parameter estimation of the Standard Model (SM) for diffusion in white matter has been elusive due to intrinsic model degeneracies and insufficient measurements. To design optimal scanner-specific protocols, we couple multidimensional protocol optimization for estimation of microstructural tissue properties in 15 minute acquisitions on clinical scanners with varying gradient performance. We show reproducible scan-rescan results and assess inter-scanner variability ranging from 1 to 8% depending on parameters. Results suggest that combining denoising, protocol optimization, and robust parameter estimation may enable quantitative microstructure mapping in clinical settings.
|
|||
3423.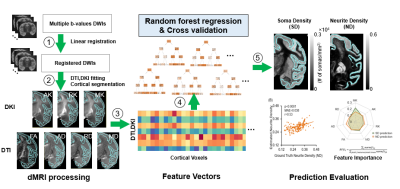 |
Estimating cortical soma and neurite densities from diffusion MRI measures using a machine learning approach
Tianjia Zhu1,2, Minhui Ouyang1, Nikou Lei3, David Wolk4, Paul Yushkevich5, and Hao Huang1,5
1Department of Radiology, Children's Hospital of Philadelphia, Philadelphia, PA, United States, 2Department of Bioengineering, University of Pennsylvania, Philadelphia, PA, United States, 3Department of Physics, University of Washington Seattle, Seattle, WA, United States, 4Department of Neurology, University of Pennsylvania, Philadelphia, PA, United States, 5Department of Radiology, University of Pennsylvania, Philadelphia, PA, United States
Diffusion MRI (dMRI) has ushered in a new era in which conventional brain cortical histological measures such as soma and neurite densities may be assessed noninvasively through advanced dMRI. However, analytical dMRI microstructural models are restricted by the model assumptions and lack of validation from quantitative histology data. Individual dMRI parameters characterize only limited microstructural information. By leveraging a variety of dMRI-based parameters delineating cortical microstructure from multiple aspects, we established a machine learning based method accurately estimating cortical soma and neurite densities in the cortex, paving the way for data-driven noninvasive virtual histology for potential applications to Alzheimer’s diseases.
|
|||
3424.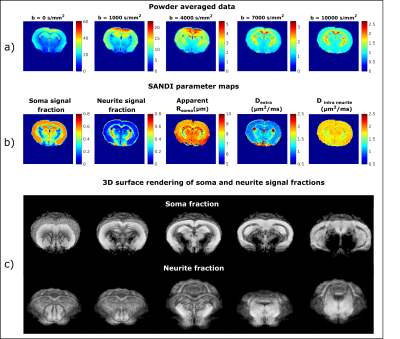 |
Mapping apparent soma and neurite density in the in-vivo mouse brain using SANDI
Andrada Ianus1, Francisca F. Fernandes1, Joana Carvalho1, Cristina Chavarrias1, Marco Palombo2, and Noam Shemesh1
1Champalimaud Centre for the Unknown, Lisbon, Portugal, 2University College London, London, United Kingdom
Measuring micro-architectural features involving cell body morphologies is emerging as a frontier of diffusion MRI. This work aimed to map the apparent soma size and density and neurite density via the SANDI methodology in the mouse brain in-vivo at 9.4T. Our results show consistent parameters values between N=3 animals in both gray and white matter ROIs. Moreover, we have also compared the effects of using different number of shells and maximum b-values in the SANDI analysis. Our work augurs well for future investigations in animal models of plasticity and disease.
|
|||
3425.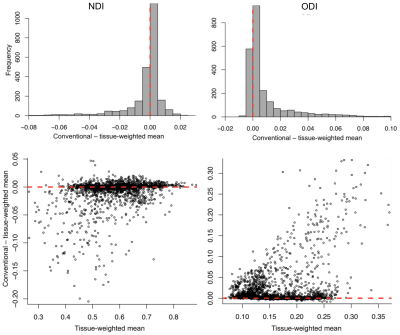 |
Not all voxels are created equal: reducing estimation bias in regional NODDI metrics using tissue-weighted mean
Christopher S Parker1, Thomas Veale2, Martina Bocchetta2, Catherine F Slattery2, Nick Fox2, Jonathan M Schott2, Dave M Cash1,2, and Hui Zhang1
1Centre for Medical Image Computing, Department of Computer Science, UCL, London, United Kingdom, 2Dementia Research Centre, Department of Neurodegenerative Disease, UCL Queen Square, Institute of Neurology, UCL, London, United Kingdom
Region-of-interest (ROI) metrics are typically computed as a mean over ROI voxels. However, for some NODDI metrics, this approach produces biased estimates in the presence of cerebrospinal fluid partial volume. We address this by introducing a tissue-weighted alternative. We compare the proposed mean to its conventional counterpart for periventricular and non-periventricular ROIs in healthy subjects and patients with young onset Alzheimer’s disease (YOAD). Results show the conventional mean overestimates orientation dispersion index and inflates inter-subject variation, particularly for periventricular ROIs and the YOAD cohort. This technique may improve detection of true regional effects in future group studies of neurodegenerative diseases.
|
|||
3426.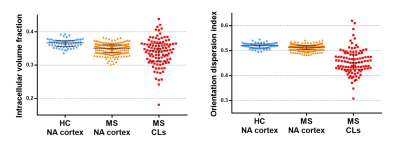 |
Relevance of NODDI to Characterise In Vivo the Microstructural Abnormalities of Multiple Sclerosis Cortex and Cortical Lesions: A 3T Study
Elisabetta Pagani1, Paolo Preziosa1,2, Raffaello Bonacchi1,2, Laura Cacciaguerra1,2,3, Massimo Filippi1,2,3,4,5, and Maria A. Rocca1,2,3
1Neuroimaging Research Unit, Division of Neuroscience, IRCCS San Raffaele Scientific Institute, Milan, Italy, 2Neurology Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 3Vita-Salute San Raffaele Unversity, Milan, Italy, 4Neurorehabilitation Unit, IRCCS San Raffaele Scientific Institute, Milan, Italy, 5Neurophysiology Service, IRCCS San Raffaele Scientific Institute, Milan, Italy
In multiple sclerosis (MS), cortical damage is a relevant predictor of clinical disability. We applied Neurite orientation dispersion and density imaging (NODDI) to characterize microstructure of normal-appearing cortex (NA-cortex) and cortical lesions (CLs) and their relations with disease clinical phenotypes and disability. We found that a significant neurite loss occurs in MS NA-cortex, being more severe with longer disease duration, more severe disability and progressive MS. CLs show a further reduction of neurite density, together with an increased extracellular space, possibly due to inflammation and gliosis, and a reduced dispersion suggestive of increased tissue coherence and simplification of neurite complexity.
|
|||
3427.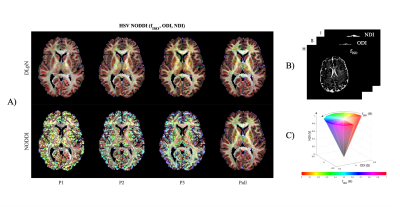 |
Deep Learner estimated isotropic volume fraction enables reliable single-shell NODDI reconstruction
Abrar Faiyaz1, Marvin M Doyley1,2,3, Giovanni Schifitto2,4, Jianhui Zhong2,3,5, and Md Nasir Uddin4
1Department of Electrical and Computer Engineering, University of Rochester, Rochester, NY, United States, 2Department of Imaging Sciences, University of Rochester, Rochester, NY, United States, 3Department of Biomedical Engineering, University of Rochester, Rochester, NY, United States, 4Department of Neurology, University of Rochester, Rochester, NY, United States, 5Department of Physics and Astronomy, University of Rochester, Rochester, NY, United States
The study explores the possibility to create neurite orientation dispersion and density imaging (NODDI) parameter maps with single-shell diffusion MRI data, using isotropic volume fraction (fISO) as prior. Proposed dictionary based deep learning prior NODDI (DLpN) framework leverages fISO estimation with a diffusion imaging scalar and T2 weighted non-diffusion signal. fISO estimation was validated in simulation and in-vivo. DLpN derived NDI and ODI maps for single-shell protocols are comparable with original multi-shell NODDI (error <5%) and may allow NODDI evaluation of retrospective brain studies on single-shell diffusion MRI data by multi-shell scanning of two subjects for DictNet fISO training.
|
|||
3428.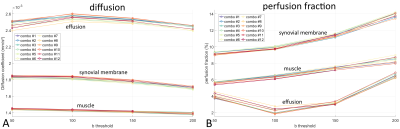 |
B-value influence on IVIM MRI for juvenile idiopathic arthritis in the knee
Kilian Stumpf1, Anna-Katinka Bracher2, Thomas Hüfken1, Britta Huch2, Meinrad Beer2, Henning Neubauer2, and Volker Rasche1
1Department of Internal Medicine II, Ulm University Medical Center, Ulm, Germany, 2Department of Diagnostic and Interventional Radiology, Ulm University Medical Center, Ulm, Germany
Intravoxel incoherent motion imaging allows the quantification of diffusion and pseudo-perfusion in tissues and can be used as an alternative to conventional T1-weighted scans that require the use of contrast agents. Its benefits for diagnosing juvenile idiopathic arthritis(JIA) in the knee has previously be demonstrated, but usually requires multi-b-value DWI scans with long scan times. In this work we investigate the influence of different b-value combinations on the calculated diffusion and perfusion fraction values and explore the possibility for IVIM MRI of JIA with fewer b-values and shorter scan times.
|
|||
3429.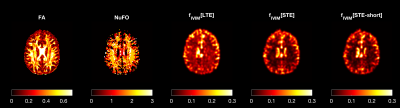 |
Fast and accurate quantification of intra-voxel incoherent motion (IVIM) with spherical-tensor-encoded diffusion MRI
Alberto De Luca1,2, Geert-Jan Biessels1, Ofer Pasternak3, and Chantal MW Tax4,5
1Department of Neurology, University Medical Center Utrecht, Utrecht, Netherlands, 2PROVIDI Lab, Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 3Brigham and Women's Hospital, Harvard Medical School, Boston, MA, United States, 4Cardiff University Brain Research, Imaging Centre (CUBRIC), University of Cardiff, Cardiff, United Kingdom, 5Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands
Diffusion MRI scans sensitive to intra-voxel incoherent motion (IVIM) are increasingly being applied to derive properties of capillary blood pseudo-diffusion in-vivo. A robust quantification of pseudo-diffusion measures requires the acquisition of diffusion MRI with multiple weightings and directions, which is time-consuming. Here, we investigate whether the isotropic averaging of spherical tensor diffusion encoding (STE) can improve IVIM estimates and shorten the acquisition time. Our simulations and in-vivo data show that STE can substantially shorten the acquisition time required to robustly quantify pseudo-diffusion while also removing a previously unreported bias in voxels containing complex white matter fiber configurations.
|
|||
3430.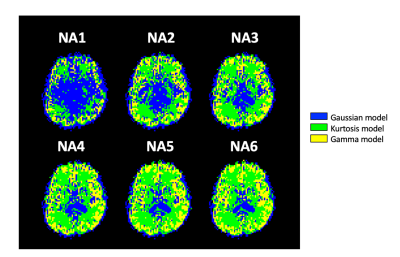 |
Effect of the Signal-to-noise Ratio on the Optimal Model Mapping for Intravoxel Incoherent Motion MRI
Yen-Peng Liao1,2, Shin-ichi Urayama1,2, Tadashi Isa1,2,3, and Hidenao Fukuyama2,4
1Department of Neuroscience, Kyoto University Graduate School of Medicine, Kyoto, Japan, 2Human Brain Research Center, Kyoto University Graduate School of Medicine, Kyoto, Japan, 3Institute for the Advanced Study of Human Biology (ASHBi), Kyoto University, Kyoto, Japan, 4Yasu City Hospital, Yasu, Japan
To improve the uncertainties caused by the inappropriate diffusion model fitting in the IVIM-MRI, an optimal model mapping method has been proposed and demonstrated its usefulness. Here, we investigated the effect of the SNR in the method. Six different SNR data sets, derived by changing the numbers of average (NA) of six sets of DWI volumes, were analyzed. Then, the resultant maps reached a steady-state when the NA is four or higher. This study revealed that further investigations on b-value distribution optimization and denoising are necessary to obtain such high SNR data within practical scan time.
|
|||
3431.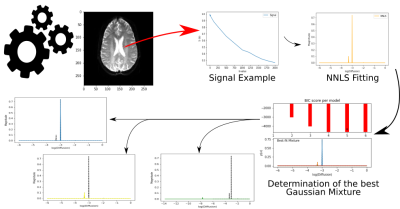 |
Gaussian Mixture for Peak Identification in Non-Negative Least Squares Fitting of the IVIM Signal
Lucas M da Costa1, Bruno Hebling Vieira1, Renata Ferranti Leoni1, and Andre Monteiro Paschoal1,2
1InBrain Lab - University of Sao Paulo, Ribeirao Preto, Brazil, 2LIM44, Instituto e Departamento de Radiologia, Faculdade de Medicina, Universidade de Sao Paulo, Sao Paulo, Brazil
The analysis of the Intravoxel Incoherent Motion (IVIM) signal can be performed with the Non-Negative Least Squares (NNLS) method. It allows detecting an indeterminate number of peaks associated with the multiple exponentials that compose the IVIM signal. Thus, it becomes essential to identify and classify these peaks. We performed a new method to analyze the NNLS spectrum applying the Gaussian Mixture. We obtained pseudo-diffusion maps with better contrast to visualize gliomas. Gaussian Mixture seems to replace the traditional peak search and classification algorithms, opening up possibilities to explore the NNLS spectrum.
|
|||
3432.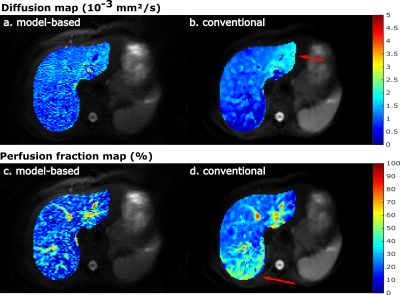 |
Model-based reconstruction for IVIM and combined IVIM-DTI fitting: Initial experience
Susanne Rauh1, Oliver Maier2, Oliver Gurney-Champion3, Melissa Hooijmans3, Rudolf Stollberger2,4, Aart Nederveen3, and Gustav Strijkers1
1Department of Biomedical Engineering and Physics, Amsterdam UMC, location AMC, Amsterdam, Netherlands, 2Institute of Medical Engineering, Graz University of Technology, Graz, Austria, 3Department of Radiology and Nuclear Medicine, Amsterdam UMC, location AMC, Amsterdam, Netherlands, 4BioTechMed-Graz, Graz, Austria
Model-based reconstruction can overcome many short-comings of IVIM and DTI by accelerating the acquisition, suppressing systematic errors from Rician noise and improving SNR. In this study we assess the feasibility of model-based reconstruction to obtain IVIM and combined IVIM-DTI parameter maps in the liver and kidneys in healthy volunteers. Results were compared to a conventional IVIM and IVIM-DTI least-squares fitting. Model-based reconstructions produced maps with less artifacts in liver and more details in the kidneys as compared to those from conventional fitting. Mean parameter values were similar for both methods.
|
|||
3433.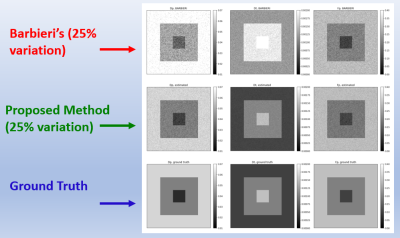 |
Physically Motivated Deep-Neural Networks of the Intravoxel Incoherent Motion Signal Decay Model for Quantitative Diffusion-Weighted MRI
Shira Nemirovsky-Rotman1, Elad Rotman1, Onur Afacan2, Sila Kurugol2, Simon Warfield2, and Moti Freiman1
1Biomedical Engineering, Technion, Haifa, Israel, 2Boston's Children's Hospital, Harvard Medical School, Boston, MA, United States
Quantitative Diffusion-Weighted MRI with the Intra-Voxel Incoherent Motion (IVIM) model shows potential to produce quantitative biomarkers for multiple clinical applications. Recently, deep-learning (DL) models were proposed for the estimation of the IVIM model parameters from DW-MRI data. While the DL models produce more accurate parameter estimates compared to classical methods, their capability to generalize the IVIM model to different acquisition protocols is very limited. For that end, we introduce a physically motivated DL model, by incorporating the acquisition protocol into the network architecture. Our approach provides a DL-based method for IVIM parameter estimates that is agnostic to the acquisition protocol.
|
|||
3434.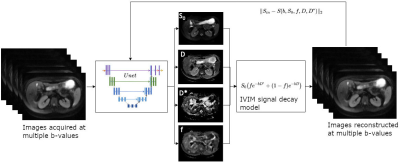 |
Self-supervised IVIM DWI parameter estimation with a physics based forward model
Serge Vasylechko1,2, Simon K. Warfield1,2, Onur Afacan1,2, and Sila Kurugol1,2
1Computational Radiology Laboratory, Boston Children's Hospital, Boston, MA, United States, 2Harvard Medical School, Boston, MA, United States
The goal of this study was to assess the robustness and repeatability of intravoxel incoherent motion model (IVIM) parameter estimation for the diffusion weighted MRI in the abdominal organs under the constraints of noisy diffusion signal using a novel neural network training method. The method is based on the principle of a physics guided self-supervised neural network that does not require supervision for training. Such approach is beneficial in conditions where the reference methods are not available, or are not robust enough to provide good supervision. This work is targeting evaluations towards accelerated IVIM DWI scanning which exhibit low SNR.
|
|||
3435.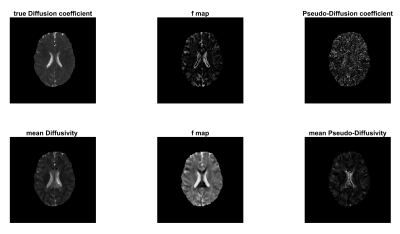 |
Intravoxel Incoherent Motion Reconstruction with Multi-Orientation Acquisition using Three b-Values
Sam Sharifzadeh Javidi1 and Hamidreza Saligheh Rad1,2
1Department of Medical Physics and Biomedical Engineering, Tehran University of Medical Sciences, Tehran, Iran (Islamic Republic of), 2Quantitative MR Imaging and Spectroscopy Group, Research Center for Molecular and Cellular Imaging, Tehran, Iran (Islamic Republic of)
The IVIM model is capable of extracting functional and structural information simultaneously without the injection of contrast agents. The main limitation of this technique is the inaccuracy of the output of this model in low SNR regimes. In this study, we proposed the use of twelve diffusion imaging orientations and three b-values instead of three orthogonal DW imaging and several b-values. Simulation and in-vivo results showed that the proposed method outperforms the conventional IVIM reconstruction method. Improved quality and reproducibility can make this method more practical and attractive in clinical settings.
|
|||
3436.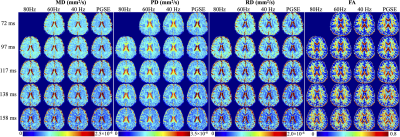 |
Time-dependent and TE-dependent Diffusivity in Human Brain using Multi-TE Oscillating Gradient Spin Echo in High Gradient 3.0T MRI (MAGNUS)
Ante Zhu1, Luca Marinelli1, and Thomas K.F. Foo1
1GE Global Research, Niskayuna, NY, United States
Oscillating gradient spin echo (OGSE) diffusion imaging achieves a short diffusion time and has been performed to assess tissue characteristics at short length scale, including the correlation length of the brain tissues which may provide information on axonal beading and swelling in disease. Time-dependent diffusivity has been measured from OGSE acquisition at a fixed TE in a high-performance head-only high-gradient 3.0T MRI scanner (MAGNUS). In this study, we characterized time-dependent diffusivity at varying frequencies (0~80 Hz) and TEs (72~158 ms) using multi-TE OGSE acquisition in healthy volunteers. Preliminary results showed increased mean/parallel diffusivities in corpus callosum at shorter TE.
|
|||
3437.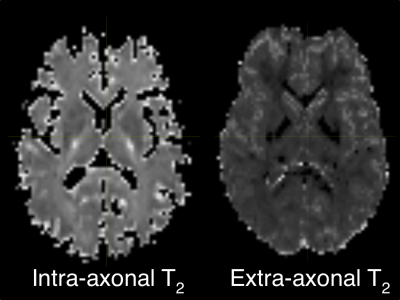 |
A novel clinically viable method to quantify T2 of intra and extra axonal compartmental tissue properties
Sudhir Kumar Pathak1, Vishwesh Nath2, B V Rathish Kumar3, and Walter Schneider1
1Psychology, University of Pittsburgh, Pittsburgh, PA, United States, 2Nvidia, Bethesda, MD, United States, 3Mathematics, Indian Institute of Technology Kanpur, Kanpur, India
Diffusion MRI based microstructural imaging is used to estimate voxel wise intra/extra-cellular volume fraction and compartment diffusivities. Previously proposed techniques like NODDI, SMT models are used to estimate intra/extra-axonal volume fraction, axial and radial diffusivity for each voxel. TEdDI is a framework to estimate intra/extra-axonal T2 in human brain. The proposed TEdDI model requires multiple sets of diffusion dataset varies with echo time. In this study we are proposing SMT based formulation to estimate intra/extra-axonalT2. We also used this formulation to design numerical algorithm that approximate T2 and other parameters for a typical multi-shell diffusion acquisition.
|
|||
3438.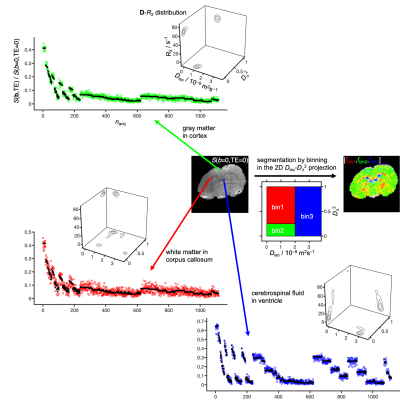 |
Nonparametric 5D D-R2 distribution imaging with single-shot EPI at 21.1 T: Initial results for in vivo rat brain
Jens T Rosenberg1, Samuel Colles Grant1,2, and Daniel Topgaard3
1National High Magnetic Field Laboratory, Florida State University, Tallahassee, FL, United States, 2Chemical and Biomedical Engineering, FAMU-FSU College of Engineering, Tallahassee, FL, United States, 3Physical Chemistry, Lund University, Lund, Sweden
In vivo diffusion MRI is by default performed using single-shot EPI with TE>50 ms and associated signal loss from transverse relaxation. The individual benefits of the current trends of increasing B0 to boost SNR and employing more advanced signal preparation schemes to improve the specificity for selected microstructural properties eventually may be cancelled by increases in relaxation rates at high B0 and TE with advanced encoding. Here, we make initial attempts to translate state-of-the-art diffusion-relaxation correlation methods to 21.1T to identify hurdles that need to be overcome to fulfill the promises of both high SNR and readily interpretable microstructural information.
|
|||
3439.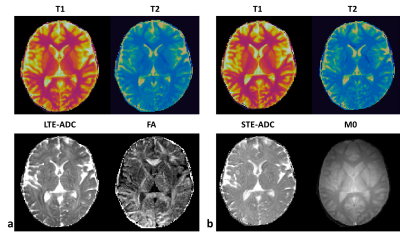 |
MR Fingerprinting with B-tensor encoding scheme for simultaneous measure of relaxation and microstructure diffusion
Maryam Afzali1, Lars Mueller1, Ken Sakaie2, Siyuan Hu3, Yong Chen4, Mark Griswold4, Derek K Jones1, and Dan Ma3
1Cardiff University Brain Research Imaging Centre (CUBRIC), School of Psychology, Cardiff University, Cardiff, United Kingdom, 2Imaging Institute, Cleveland Clinic, Cleveland, OH, United States, 3Biomedical Engineering, Case Western Reserve University, Cleveland, OH, United States, 4Radiology, Case Western Reserve University, Cleveland, OH, United States
Diffusion magnetic resonance imaging provides information about microstructural features of a sample. Relaxometry on the other hand is sensitive to the biochemical environment of the underlying microstructure. Recent studies show that using conventional Stejskal-Tanner experiment, separating compartmental features is challenging and therefore b-tensor encoding and diffusion-relaxometry were proposed to mitigate this challenge. Magnetic resonance fingerprinting (MRF) can quantify multiple tissue parameters in one scan. Here, we implement multi-dimensional MR Fingerprinting scan with linear and spherical tensor encoding (LTE, and STE) and show the feasibility of estimating $$$T_1,\;\rm{and}\;T_2$$$ relaxation times and diffusivity on NIST, microstructure phantom and in vivo brain scans.
|
|||
3440.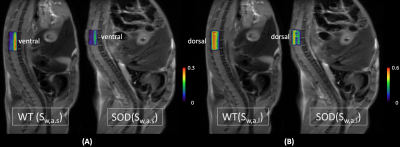 |
Multicomponent Diffusion Analysis using L1-norm Regularized NNLS for an Accurate and Robust Detection of Alternations in Spinal Cord
Jin Gao1,2, Weiguo Li2,3, Richard Magin3, and Danilo Erricolo1,3
1Department of Electrical and Computer Engineering, University of Illinois at Chicago, Chicago, IL, United States, 2Research Resources Center, University of Illinois at Chicago, Chicago, IL, United States, 3Department of Bioengineering, University of Illinois at Chicago, Chicago, IL, United States
Multiple components analysis of nuclear magnetic resonance (MR) relaxation data using L2-norm regularized non-negative least squares (NNLS) method has been widely used in myelin imaging for neurological diseases. When this analysis is applied to diffusion-weighted MR imaging to investigate water diffusion properties of biological tissues, noise corruption becomes a major problem which affects the accuracy and robustness of results. In this study, a L1-norm regularized method was developed to process diffusion-weighted MRI data from spinal cords of amyotrophic lateral sclerosis affected mice.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.