Digital Posters
Diffusion: Encoding & Estimation
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 3 | 19:00 - 20:00 |
2457.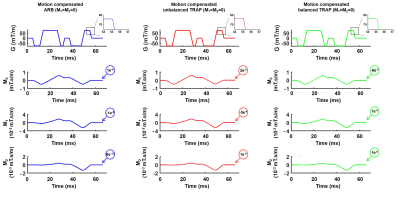 |
Benefits of arbitrary gradient waveform design for diffusion encoding
Kevin Moulin1,2,3, Mike Loecher1,2, Matthew J Middione1,2, and Daniel B Ennis1,2,3
1Department of Radiology, Stanford University, Stanford, CA, United States, 2Department of Radiology, Veterans Administration Health Care System, Palo Alto, CA, United States, 3Cardiovascular Institute, Stanford University, Stanford, CA, United States
Traditional diffusion encoding waveforms are usually composed of two symmetric trapezoidal gradients (TRAP). Shape-free arbitrary (ARB) gradient waveforms offer a higher b-value than ARBs. They can be designed analytically and symmetric or numerically and asymmetric. The objectives of this work were to analyze the performances of ARB and TRAP for asymmetric numerically designs and for symmetric analytically designed diffusion encoding gradient waveforms.
|
|||
2458.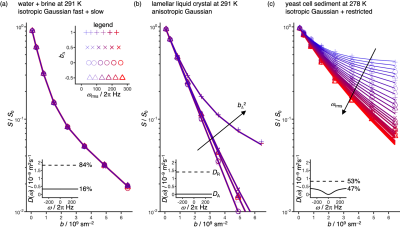 |
Gradient waveforms for comprehensive sampling of the frequency and "shape" dimensions in b(ω)-encoded diffusion MRI
Hong Jiang1 and Daniel Topgaard1
1Physical Chemistry, Lund University, Lund, Sweden
Diffusion encoding with either oscillating gradients or as a function of the "shape" of the b-tensor have both recently found powerful applications for characterization of tissue microstructure in clinical MRI. We here propose a simple scheme based on the "double rotation" technique in solid-state NMR spectroscopy to generate a family of gradient waveforms enabling comprehensive sampling of the multidimensional space defined by the tensor-valued encoding spectrum with special emphasis on the frequency and shape dimensions. The approach is demonstrated by microimaging experiments on phantoms with well-defined anisotropy and restriction properties, thereby paving the way for future clinical implementations.
|
|||
2459.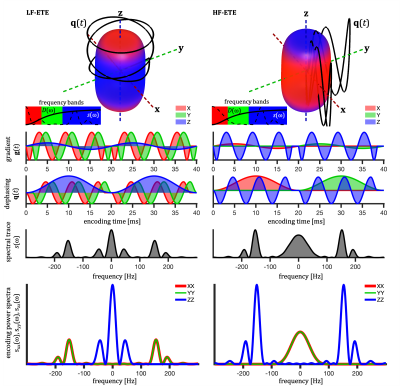 |
Tissue microstructure by ellipsoidal tensor encoding with independently varying spectral anisotropy and tuning
Samo Lasic1,2 and Henrik Lundell1
1Danish Research Centre for Magnetic Resonance, Centre for Functional and Diagnostic Imaging and Research, Copenhagen University Hospital Hvidovre, Copenhagen, Denmark, 2Random Walk Imaging, Lund, Sweden
Ellipsoidal tensor encoding (ETE) with independent control of spectral anisotropy (SA) and tuning provides two distinct encoding frequency windows in a single experiment and yields distinctly different signal signatures for compartments with different anisotropic time-dependent diffusion. ETE can be orientation invariant, depending on SA and restriction geometry. Signal orientation variation minima depends on size relative to tuning but not on orientation dispersion. This popery could be useful for quick size estimation and geometry detection. Such encoding strategy could potentially provide new contrasts sensitive to specific pathological variations.
|
|||
2460.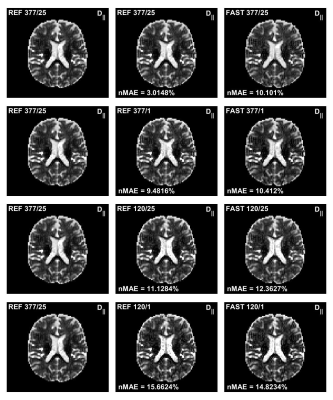 |
A Novel Fast Quantitative Parameter Distribution Estimator Applied to Diffusion Tensor Distribution Imaging
Anders Garpebring1
1Radiation Sciences, div. Radiation Physics, Umeå University, Umeå, Sweden
Non-parametric diffusion tensor distribution estimation is very computationally expensive and can require several hours of processing for a single 3D volume. In this work a new formulation of the estimation problem is developed as well as a new more efficient algorithm. The results show that the computational times can be reduced to minutes or even seconds rather than hours. Thus, making these types of analysis suitable also in a clinical setting.
|
|||
2461.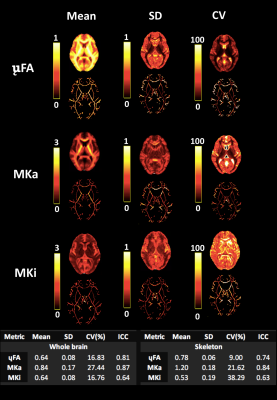 |
Quantifying the Repeatability of Microstructural Measures Derived from Free Gradient Waveforms
Kristin Koller1, Chantal MW Tax1,2, Dmitri Sastin1,3,4, and Derek K Jones1,5
1Cardiff University Brain Research Imaging Centre, Cardiff, United Kingdom, 2Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 3Department of Neurosurgery, University Hospital of Wales, Cardiff, United Kingdom, 4BRAIN Biomedical Research Unit, Health & Care Research Wales, Cardiff, United Kingdom, 5Mary MacKillop Institute for Health Research, Australian Catholic University, Melbourne, Australia
Diffusional variance decomposition (DIVIDE) is a promising state-of-the-art approach to measure microscopic microstructure using ‘free’ gradient waveforms (including spherical and linear tensor encoding). Here, in a cohort of 6 participants scanned 5 times each with the DIVIDE protocol, we quantify the microscopic FA (μFA) and isotropic and anisotropic diffusional variance (MKi and MKa) across scans, thus demonstrating high test-retest repeatability.
|
|||
2462.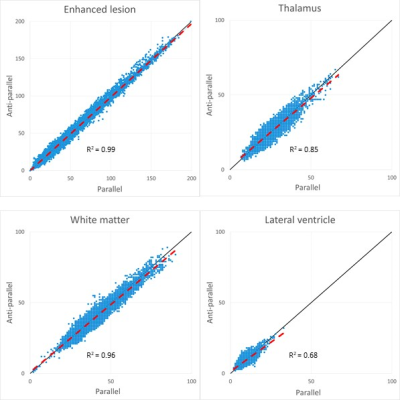 |
Adequate mixing time for double diffusion encoding in normal brain structures and brain tumors
Kentaro Akazawa1, Koji Sakai1, Tomoaki Kitaguchi1, Tomonori Toyotsuji1, Thorsten Feiweier 2, Hiroshi Imai3, and Kei Yamada1
1Radiology, Kyoto Prefectural University of Medicine, Kyoto, Japan, 2Siemens Healthcare GmbH, Erlangen, Germany, 3Siemens Healthcare K.K., Shinagawa, Japan
The mixing time for double diffusion encoding (DDE) should be set low to reduce the relaxation effects but also high enough for estimating microscopic fractional anisotropy. We tested the adequacy of the mixing time of 30 msec by comparing acquisitions with parallel and anti-parallel directions as well as with orthogonal and collinear directions. Relatively short mixing time for our cohort was adequate to evaluate the microscopic fractional anisotropy not only in the normal brain area of the white matter and the central gray matter, but also in pathologically abnormal areas such as brain tumors.
|
|||
2463.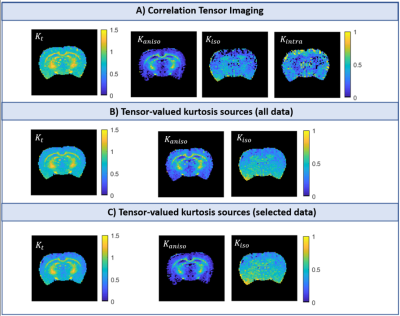 |
Intra-compartmental kurtosis biases tensor-valued multidimensional diffusion
Rafael Neto Henriques1, Sune Nørhøj Jespersen2,3, and Noam Shemesh1
1Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 2Center of Functionally Integrative Neuroscience (CFIN) and MINDLab, Clinical Institute, Aarhus University, Aarhus, Denmark, 3Department of Physics and Astronomy, Aarhus University, Aarhus, Denmark
Multidimensional diffusion encoding (MDE) has been gaining attention for its potential to describe tissue microstructure with enhanced specificity by resolving kurtosis sources, albeit with significant assumptions (no time dependence, no intra-compartmental kurtosis). Correlation Tensor Imaging (CTI) has been introduced as a novel methodology capable of resolving kurtosis sources without relying on a-priori assumptions. Here, we harnessed CTI to validate the accuracy of tensor-valued MDE metrics and assess the importance of intra-compartmental kurtosis (Kintra) in tissues. Our results reveal that Kintra is non-negligible and skews the estimates of tensor-valued MDE approaches, even in the absence of detectable diffusion time dependence.
|
|||
2464.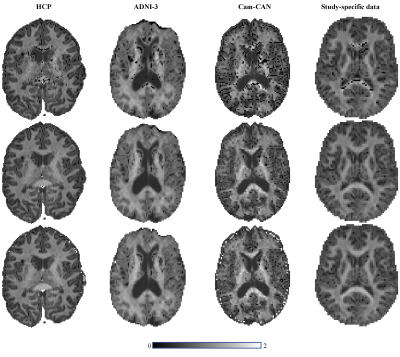 |
Towards more robust and reproducible Diffusion Kurtosis Imaging
Rafael N Henriques1, Sune N. Jespersen2,3, Derek K. Jones4,5, and Jelle Veraart6
1Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 2Department of Clinical Medicine, Aarhus University, Aarhus, Denmark, 3Department of Physics and Astronomy, Aarhus University, Aarhus, Denmark, 4School of Psychology, Cardiff University, Cardiff, United Kingdom, 5Mary MacKillop Institute for Health Research, Australian Catholic University, Melbourne, Australia, 6Center for Biomedical Imaging, NYU Grossman School of Medicine, New York, NY, United States
The general utility of Diffusion Kurtosis Imaging (DKI) is challenged by its poor robustness to imaging artifacts and thermal noise that often lead to implausible kurtosis values. A robust scalar kurtosis index can be estimated from powder-averaged diffusion-weighted data. We introduce a novel DKI estimator that uses this scalar kurtosis index as a proxy for the mean kurtosis to regularize the fit. The regularized DKI estimator improves the robustness and reproducibility of the kurtosis metrics and results in parameter maps with enhanced quality and contrast; thereby promoting the wider use of DKI in clinical research and potentially diagnostics.
|
|||
2465.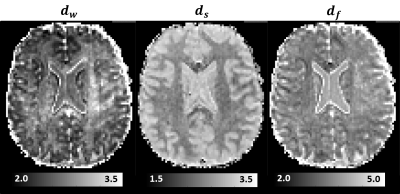 |
The diffusion time dependence of MAP-MRI parameters in the human brain
Alexandru V Avram1,2, Qiyuan Tian3, Qiuyun Fan3, Susie Y Huang3,4, and Peter J Basser1
1Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, MD, United States, 2Center for Neuroscience and Regenerative Medicine, The Henry Jackson Foundation, Bethesda, MD, United States, 3Athinoula A. Martinos Center for Biomedical Imaging, Department of Radiology, Massachusetts General Hospital, Harvard Medical School, Charlestown, MA, United States, 4Harvard-MIT Division of Health Sciences and Technology, Massachusetts Institute of Technology, Cambridge, MA, United States
We acquire mean apparent propagator (MAP) MRI data in the human brain using different diffusion times. We characterize the diffusion-time dependence of propagator metrics in vivo and compute the statistical distance between co-registered propagators measured with short and long diffusion times. We derive time-scaling parameters that can assess anomalous diffusion in disordered, fractal-like tissue environments. Preliminary results suggest that the diffusion-time dependence of in vivo MRI signals is strongly modulated by restrictions and hindrances that occur over a range of length scales and could provide new contrasts to quantify structural and architectural differences in healthy and diseased tissues.
|
|||
2466.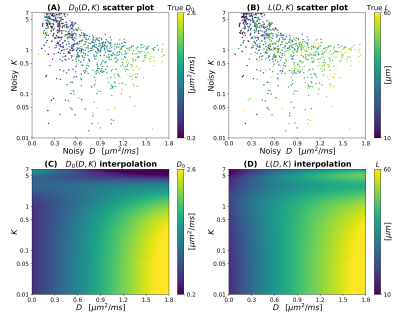 |
Investigating the relationship between diffusion MRI signal cumulants and hepatocyte microstructure at fixed diffusion time
Francesco Grussu1, Kinga Bernatowicz1, Ignasi Barba2, and Raquel Perez-Lopez1,3
1Radiomics Group, Vall d'Hebron Institute of Oncology, Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain, 2NMR Lab, Vall d'Hebron Institute of Oncology, Vall d'Hebron Barcelona Hospital Campus, Barcelona, Spain, 3Department of Radiology, Hospital Universitari Vall d'Hebron, Barcelona, Spain
To date, limited attention has been paid to diffusion-weighted (DW) MRI signal modelling of the liver, where new imaging methods are needed to tackle diseases such as cancer. We report on Monte Carlo (MC) simulations run in synthetic hepatic cells to inform the developing of new model-based methods for liver application. We specifically investigate the question: “can cell size and diffusivity be estimated from signal cumulants at fixed diffusion time and realistic SNR?”, and find that the task is feasible for clinical diffusion times and b=0 SNR as low as 20, provided that both apparent diffusivity and kurtosis are considered.
|
|||
2467.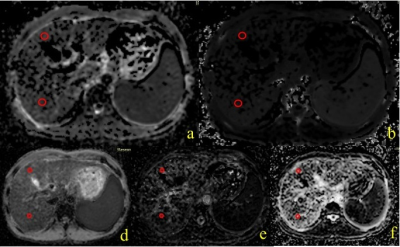 |
Application of DKI and IVIM in staging of hepatic fibrosis
YANLI JIANG1, Jie Zou1, FengXian Fan1, Yuping Bai1, Jing Zhang1, and Shaoyu Wang2
1Department of Magnetic Resonance, LanZhou University Second Hospital, LanZhou, China, 2MR Scientific Marketing, Siemens Healthineers, Shanghai, China
Intravoxel Incoherent Motion (IVIM) is a MRI method that enables simultaneous assessment of diffusion and perfusion. Diffusion Kurtosis imaging (DKI) is based on a mathematical approach that uses a polynomial model with a dimensionless factor called the kurtosis (K). This study evaluated the relationship between IVIM-DKI diffusion models and clinical-pathologic to assess their diagnostic accuracy for staging of hepatic fibrosis. We found that a high level of hepatic fibrosis usually have a higher MK values. A significant negative correlation was observed between FibroScan and MD. In conclusion, compared with IVIM, DKI might be more useful in staging of hepatic fibrosis.
|
|||
2468.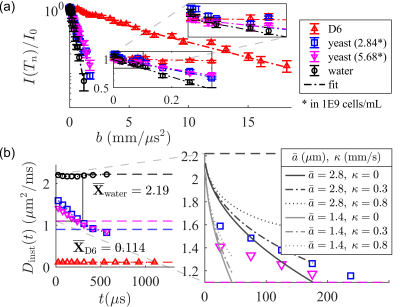 |
Single-shot measurement of sub-millisecond, time-dependent diffusion using optimized, unequal pulse spacings in a static field gradient
Teddy Xuke Cai1,2, Nathan Hu Williamson1,3, Velencia Witherspoon1, Rea Ravin1,4, and Peter Basser1
1Section on Quantitative Imaging and Tissue Sciences, Eunice Kennedy Shriver National Institute of Child Health and Human Development, Bethesda, MD, United States, 2Wellcome Centre for Integrative Neuroimaging, University of Oxford, Oxford, United Kingdom, 3National Institute of General Medical Sciences, Bethesda, MD, United States, 4Celoptics, Inc., Rockville, MD, United States
Time-dependent diffusion contains rich information about the tissue microstructure. Conventional methods to measure the time-varying diffusivity probe a single timescale per acquisition, limiting time resolution. Furthermore, access to sub-millisecond timescales is limited by the pulsed gradient hardware. An alternative method is presented here. We extend the static field gradient, Carr-Purcell-Meiboom-Gill cycle by incrementing the $$$\pi$$$-pulse spacings to isolate the on-resonance signal. The resulting spin echo train probes a range of short timescales (50 – 500 microseconds) in one shot and enables a 1-minute time-dependent diffusivity measurement. Proof-of-principle simulations and experimental results on pure liquids and yeast are presented.
|
|||
2469.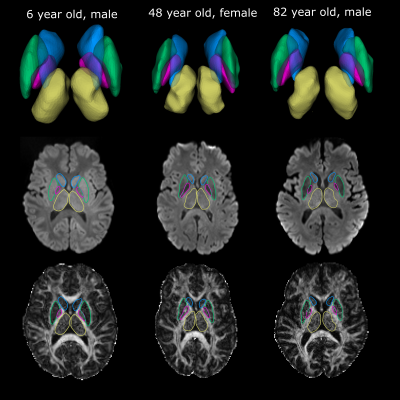 |
Automated Surface-Based Segmentation of Deep Grey Matter Brain Regions Based Solely on Diffusion Tensor Images
Graham Little1 and Christian Beaulieu1
1Biomedical Engineering, University of Alberta, Edmonton, AB, Canada
A surface-based deep grey matter segmentation algorithm is proposed that works directly on diffusion images and maps of the brain acquired at a 1.5 mm isotropic resolution. The method was applied to twenty participants spanning a large age range (6-90 years) resulting in accurate segmentations of the thalamus, caudate, putamen and globus pallidus. Fractional anisotropy and mean diffusivity showed unique non-linear trajectories across the lifespan. The proposed method avoids the need of problematic coregistration to other scans (anatomical T1) and will accelerate the analysis of microstructural changes of deep grey matter regions with age (or disease) in large populations.
|
|||
2470.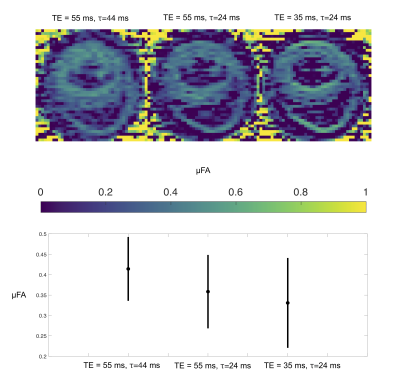 |
Investigating time dependent diffusion, microscopic anisotropy and T2 effects in the mouse heart
Henrik Lundell1, Samo Lasič1,2, Filip Szczepankiewicz3,4,5, Beata Wereszczyńska6, Matthew Budde7, Erica Dall'Armellina6, Nadira Yuldasheva6, Jürgen E. Schneider6, and Irvin Teh6
1Danish Research Centre for Magnetic Resonance, Centre for Functional and Diagnostic Imaging and Research, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark, 2Random Walk Imaging, Lund, Sweden, 3Clinical Sciences, Lund University, Lund, Sweden, 4Harvard Medical School, Boston, MA, United States, 5Brigham and Women's Hospital, Boston, MA, United States, 6Leeds Institute of Cardiovascular and Metabolic Medicine, University of Leeds, Leeds, United Kingdom, 7Department of Neurosurgery, Medical College of Wisconsin, Milwaukee, WI, United States
Non-invasive characterization of cardiac microstructure by diffusion MRI has provided insights into the healthy and diseased heart. Multidimensional diffusion encoding (MDE) aims for measurements with independent contrasts for specific effects. We suggest a battery of MDE measurements that probe diffusivity and microscopic anisotropy at different diffusion and echo times. We show a clear effect of time-dependent diffusion but a smaller effect from transversal relaxation.
|
|||
2471.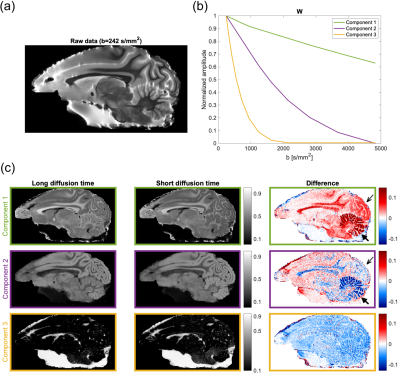 |
Data-driven separation of MRI signal components for tissue characterization
Sofie Rahbek1, Kristoffer H. Madsen2,3, Henrik Lundell2, Faisal Mahmood4,5, and Lars G. Hanson1,2
1Department of Health Technology, Technical University of Denmark, Kgs. Lyngby, Denmark, 2Danish Research Centre for Magnetic Resonance, Centre for Functional and Diagnostic Imaging and Research, Copenhagen University Hospital Hvidovre, Hvidovre, Denmark, 3Department of Applied Mathematics and Computer Science, Technical University of Denmark, Kgs. Lyngby, Denmark, 4Laboratory of Radiation Physics, Odense University Hospital, Odense, Denmark, 5Department of Clinical Research, University of Southern Denmark, Odense, Denmark
We propose a novel data-driven method for extraction of tissue-related signal components from high-dimensional MRI data. In this method, the standard non-negative matrix factorization (NMF) has been extended with signal monotonicity constraints suitable for several MR signal types, and is termed the monotonous slope NMF (msNMF). Its applications are here demonstrated using both diffusion-weighted and relaxometry data. The msNMF successfully distinguish areas with different cell densities and levels of white matter intra-myelinic edema, respectively, and is potentially useful for diagnosis and therapy evaluation.
|
|||
2472.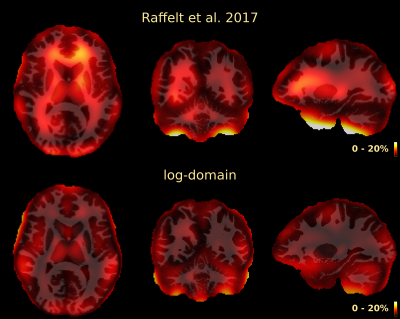 |
Multi-tissue log-domain intensity and inhomogeneity normalisation for quantitative apparent fibre density
Thijs Dhollander1,2, Rami Tabbara2, Jonas Rosnarho-Tornstrand3,4, J-Donald Tournier3,4, David Raffelt2, and Alan Connelly2
1Developmental Imaging, Murdoch Children's Research Institute, Melbourne, Australia, 2Florey Institute of Neuroscience and Mental Health, University of Melbourne, Melbourne, Australia, 3Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 4Department of Biomedical Engineering, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom Multi-tissue constrained spherical deconvolution of diffusion MRI data yields white matter fibre orientation distributions, from which a quantitative metric of apparent fibre density can be obtained. Unlike most other diffusion MRI models, this fibre density metric is directly proportional to the diffusion-weighted signal magnitude, and thus intensity normalisation and bias field correction are needed to compare it between subjects in a study. Here we propose an intensity and inhomogeneity correction algorithm for multi-tissue constrained spherical deconvolution results, estimating a bias field and global normalisation in the log-domain. It outperforms a previously proposed approach that did not operate in the log-domain. |
|||
2473.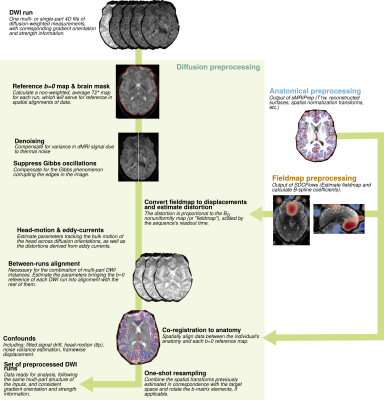 |
dMRIPrep: a robust preprocessing pipeline for diffusion MRI
Michael J Joseph1, Derek Pisner2, Adam Richie-Halford3, Garikoitz Lerma-Usabiaga4, Salim Mansour1, James D Kent5, Anisha Keshavan3, Matthew Cieslak6, Erin W Dickie1, Sebastian Tourbier7, Aristotle N Voineskos1, Theodore D Satterthwaite6, Russell A Poldrack8,
Jelle Veraart9, Ariel Rokem10, and Oscar Esteban7
1The Centre for Addiction and Mental Health, Toronto, ON, Canada, 2Department of Psychology, University of Texas at Austin, Austin, TX, United States, 3eScience Institute, The University of Washington, Seattle, WA, United States, 4Basque Center on Cognition, Brain and Language, Donostia - San Sebastian, Spain, 5Neuroscience Program, University of Iowa, Iowa City, IA, United States, 6Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 7Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 8Department of Psychology, Stanford University, Stanford, CA, United States, 9NYU Grossman School of Medicine, New York City, NY, United States, 10Department of Psychology, The University of Washington, Seattle, WA, United States
We present dMRIPrep, a preprocessing pipeline for diffusion MRI (dMRI) inspired by the approach and wide uptake of fMRIPrep. dMRIPrep reliably and consistently performs on diverse data acquired by different studies. dMRIPrep equips researchers with a reliable and transparent tool developed with the best available engineering practices and neuroimaging standards, maintained as part of NiPreps, a community software framework that ensures long-lasting support and public-interest steering.
|
|||
2474.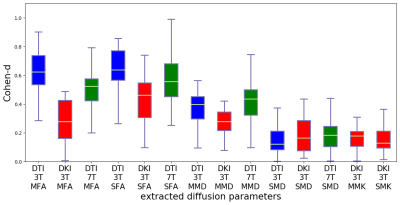 |
Sensitivity to WM injury in SLE assessed by diffusion MRI: influence of field strength, acquisition approach and post-processing strategy
Evgenios N. Kornaropoulos1,2, Stefan Winzeck2,3, Theodor Rumetshofer1, Anna Wikstrom1, Linda Knutsson4,5, Marta Correia6, Pia Sundgren1,7,8, and Markus Nilsson1
1Diagnostic Radiology, Lund University, Lund, Sweden, 2Division of Anaesthesia, University of Cambridge, Cambridge, United Kingdom, 3Department of Computing, Imperial College London, London, United Kingdom, 4Department of Medical Radiation Physics, Lund University, Lund, Sweden, 5Russell H. Morgan Department of Radiology and Radiological Science, Johns Hopkins University, Baltimore, MD, United States, 6MRC Cognition and Brain Sciences Unit, University of Cambridge, Cambridge, United Kingdom, 7Lund University BioImaging Center, Lund University, Lund, Sweden, 8Department of Medical Imaging and Physiology, Skane University Hospital, Lund, Sweden
There are many ways to acquire and process diffusion MRI data. However, it is not known which yields the largest effect sizes in group studies. We evaluated the impact of different acquisitions (3T versus 7T and DTI versus DKI) and post-processing strategies (motion correction, Gibbs-ringing and denoising) on effect size estimates of white matter (WM) injury in patients with Systemic Lupus Erythematosus (SLE). Results showed that fractional anisotropy (FA) from a 3T DTI acquisition yielded the largest effect sizes.
|
|||
2475.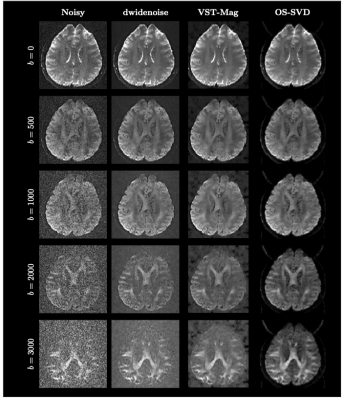 |
Reducing Noise in Complex-Valued Multi-Channel Diffusion-Weighted Data via Optimal Shrinkage of Singular Values
Khoi Minh Huynh1,2, Wei-Tang Chang2, and Pew-Thian Yap1,2
1Biomedical Engineering, UNC Chapel Hill, Chapel Hill, NC, United States, 2Department of Radiology and Biomedical Research Imaging Center (BRIC), UNC Chapel Hill, Chapel Hill, NC, United States
We evaluated the effectiveness of optimal-shrinkage singular value decomposition (OS-SVD) in denoising multi-channel complex-valued diffusion MRI data prior to image reconstruction and show that it outperforms other state-of-the-art denoising methods.
|
|||
2476.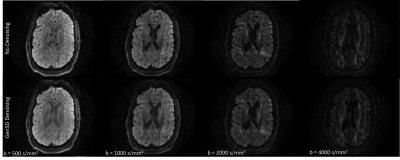 |
Model Based Denoising of Diffusion MRI Reduces Bias in Tensor Derived Parameters and Connectivity Measures
Nastaren Abad1, Luca Marinelli1, Radhika Madhavan1, and Tom K.F Foo1
1General Electric Global Research, Niskayuna, NY, United States
Higher spatial and angular resolution is essential in diffusion MRI to resolve fiber and structural ambiguities. However, quantitative measures are confounded by low SNR, particularly at high b-values, compensation of which leads to longer acquisition times. In this study, the fundamental question asked is: Can denoising aid the stability of the measurement in the presence of increasing noise? Model based denoising was used to explore accelerated sampling by evaluating bias developed in qualitative and quantitative end points. Experimental results highlight superior performance, compared to ground truth, in noise and bias reduction in metrics along with structure preservation.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.