Digital Posters
Machine Learning Applications in CV Imaging I
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 2 | 13:00 - 14:00 |
2634.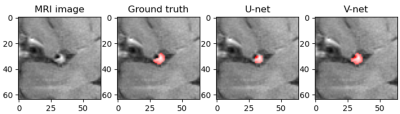 |
Automatic segmentation of middle cerebral artery plaque based on deep learning
Shuai Shen1,2,3,4, Xiao Liu5, Zhuyuerong Li5, Tao Jiang5, Hairong Zheng1,3,4, Xin Liu1,3,4, and Na Zhang1,3,4
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 2College of Software, Xinjiang University, Urumqi, China, 3Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 4CAS key laboratory of health informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 5Department of radiology, Beijing Chao-Yang hospital, Capital medical university, beijing, China
At present, deep learning has gradually been applied to the field of plaque segmentation. However, the existing work is mainly used for the processing of 2D images. In this study, we trained a 3D network model to automatically segment the middle cerebral artery plaques based on 3D images and compared the accuracy with 2D network model. Magnetic resonance vessel wall imaging (MR-VWI) data from 102 patients were used for training. The results showed that all quantitative accuracy indicators of V-net were higher than U-net, and experiments showed that V-net was more stable.
|
|||
2635.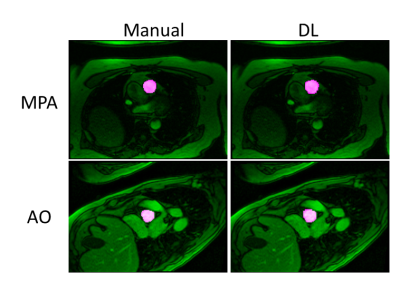 |
Automated Vessel Segmentation for 2D Phase Contrast MR Using Deep Learning
Ning Jin1, Maria Monzon2, Teodora Chitiboi3, Aaron Pruitt4, Daniel Giese2, Matthew Tong5, and Orlando P Simonetti5,6,7
1Cardiovascular MR R&D, Siemens Medical Solutions USA, Inc., Cleveland, OH, United States, 2Siemens Healthcare, Erlangen, Germany, 3Siemens Medical Solutions USA, Inc, Princeton, NJ, United States, 4Biomedical Engineering, The Ohio State University, Columbus, OH, United States, 5Internal Medicine, The Ohio State University, Columbus, OH, United States, 6Davis Heart & Lung Research Institute, The Ohio State University, Columbus, OH, United States, 7Radiology, The Ohio State University, Columbus, OH, United States
Phase-contrast (PC) MRI is used to evaluate blood hemodynamics; however, it can be time consuming to process PC-MR data. In this work, we developed a fully automated segmentation algorithm for PC MR images using deep learning (DL). Automated segmentation of aorta and main pulmonary artery from PC MRI scans can be successfully achieved using the DL model.
|
|||
2636.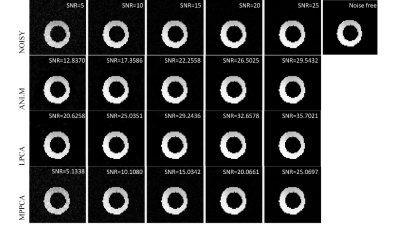 |
The Comparison of denoising methods for cardiac diffusion tensor imaging
Xi Xu1, Yuxin Yang1, Yuanyuan Liu1, Dong Liang1, Hairong Zheng1, and Yanjie Zhu1
1Shenzhen Institute of Advanced Technology, ShenZhen, China
We evaluate three different image denoising methods in cardiac diffusion tensor imaging (CDTI) regarding image quality and accuracy of parameter estimates with simulation and ex-vivo experiments. The local principal component analysis (LPCA) performs the best in improving image quality both in simulated and ex-vivo data, and the uncertainty of parameter estimations is reduced by all three algorithms in the ex-vivo experiment.
|
|||
2637.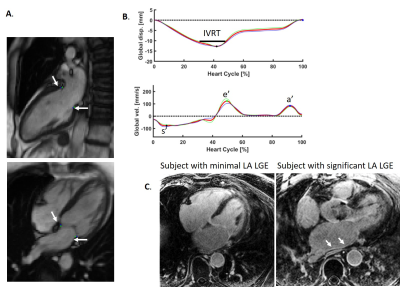 |
Isovolumic Relaxation Time and e’ Metrics Evaluated by Deep-learning Analysis of Long-axis Cine: Correlations to Atrial Pressure and Fibrosis
Dana Peters1, Jérôme Lamy1, Felicia Seemann2, Einar Heiberg3, and Ricardo Gonzales1
1Yale Unversity, New Haven, CT, United States, 2National Institutes of Health, Bethesda, MD, United States, 3Lund University, Lund, Sweden
Using a deep-learning tool for tracking the left ventricular valve plane on long-axis cine, markers of diastolic dysfunction could be easily evaluated, including e-prime, a-prime (atrial kick valve velocity), s-prime, and isovolumic relaxation time (IVRT). This analysis, performed in 23 patients with atrial fibrillation, revealed a dependence of both a-prime and IVRT on atrial fibrosis.
|
|||
2638.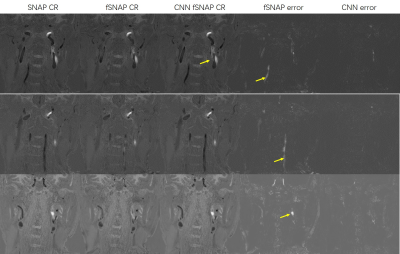 |
Comparison of Traditional fSNAP and 3D FuseUnet Based fSNAP
Chuyu Liu1, Shuo Chen1, and Rui Li1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China
By adapting 3D FuseUnet, CNN fSNAP showed better performance in lumen and IPH depiction compared with traditional fSNAP. The results suggest that deep learning can help fast SNAP scans produce high quality images, which could have great clinical utility.
|
|||
2639.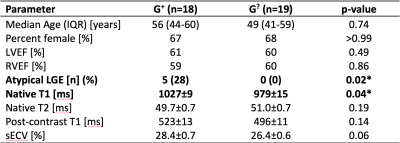 |
Deep phenotyping of individuals with arrhythmogenic cardiomyopathy-associated genetic variants using myocardial T1 and T2 mapping
Eric D Carruth1, Samuel W Fielden1, Amro Alsaid1, Brandon K Fornwalt1, and Christopher M Haggerty1
1Geisinger, Danville, PA, United States
As genomic screening initiatives expand, pre-symptomatic identification of genetic risk of disease is increasingly common. For example, Geisinger’s Genomic Screening and Counseling program has identified carriers of rare variants associated with arrhythmogenic cardiomyopathy, but most have no clinical diagnosis. We explored whether myocardial T1 or T2 maps or synthetic extracellular volume (sECV) in these patients could assist in risk stratification by identifying pre-clinical myocardial abnormalities. We compared these quantitative measures to those of controls with a normal cardiac MRI. Native T1 was significantly elevated in asymptomatic variant carriers; however, T2, post-contrast T1, and sECV values were comparable.
|
|||
2640.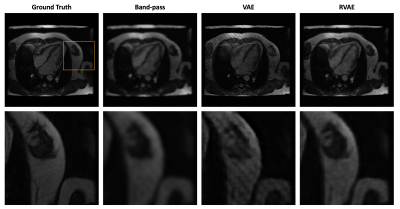 |
Unsupervised Tag Removal in Cardiac Tagged MRI using Robust Variational Autoencoder
Botian Xu1 and John C. Wood1,2
1Department of Biomedical Engineering, University of Southern California, Los Angeles, CA, United States, 2Division of Cardiology, Children's Hospital Los Angeles, Los Angeles, CA, United States
Post processing of cardiac tagged MRI has always been challenging because of poor SNR and image artifact. Inspired by unsupervised anomalies detection using variational autoencoder (VAE), we treat tags as anomalies and employ robust variational autoencoder (RVAE), using \(\beta\)-ELBO cost, which is more robust to outliers, to replace the log-likelihood optimization of a prototypical VAE, to generate tag-free results from cardiac tagged images.
|
|||
2641.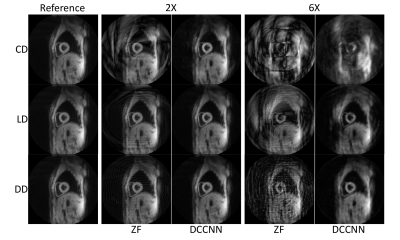 |
A comparison of spiral trajectories in a deep learning reconstruction for DENSE
Samuel Fielden1,2, Eric Carruth1, Brandon Fornwalt1,3,4, and Christopher Haggerty1,3
1Translational Data Science and Informatics, Geisinger, Danville, PA, United States, 2Medical and Health Physics, Geisinger, Danville, PA, United States, 3Heart Institute, Geisinger, Danville, PA, United States, 4Radiology, Geisinger, Danville, PA, United States
Displacement Encoding with Stimulated Echoes (DENSE) is a powerful technique that has found great utility in accurately measuring cardiac tissue displacement. However, DENSE remains time-consuming to acquire, particularly for 3-dimensionally encoded or higher resolution schemes, so methods to accelerate image acquisition are needed. Deep learning has shown promise to assist with a myriad of reconstruction problems, including DENSE. Here, we explore the reconstruction performance of a non-Cartesian Deep Cascade of Convolutional Neural Networks (DCCNN) when presented with undersampled data generated from multiple spiral trajectory designs and acceleration rates.
|
|||
2642.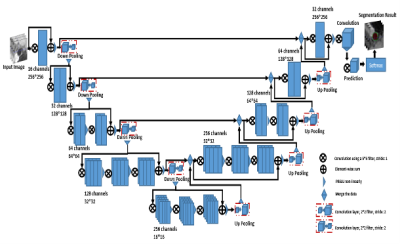 |
Deep Learning for MR Vessel Wall Imaging: Automated Detection of arterial vessel wall and plaque
Wenjing Xu1,2,3,4, Xiong Yang5, Yikang Li1,3,4, Jin Fang6, Guihua Jiang6, Shuheng Zhang5, Yanqun Teng5, Xiaomin Ren5, Lele Zhao5, Jiayu Zhu5, Qiang He5, Hairong Zheng1,3,4, Xin Liu1,3,4, and Na Zhang1,3,4
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Faculty of Information Technology, Beijing University of Technology, Beijing, China, 3Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 4CAS key laboratory of health informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 5Shanghai United Imaging Healthcare Co., Ltd., Shanghai, China, 6Department of Radiology, Guangdong Second Provincial General Hospital, Guangzhou, China
The rupture of unstable or vulnerable atherosclerotic plaque is the major cause of ischemic stroke. Manual analysis of the vessel wall and plaque is labor-intensive and experience dependent. The purpose of this study is to develop an automatic method to segment the vessel wall and lumen contour for quantitative measurement. In this work, a CNN architecture for fully automated segmentation of arterial lumen and vessel wall on MR vessel wall images was developed and evaluated on ischemic stroke patients. In conclusion, we proposed a good performance automatic analysis method for the vessel wall and is important for plaque analysis.
|
|||
2643.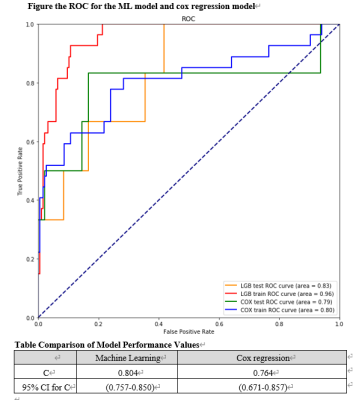 |
A Machine Learning Approach for Predicting cardiovascular event in HCM patient on Cardiac MRI
kankan hao1,2, yanjie zhu1,2, dong liang1,2, shihua zhao3, xin liu1,2, and hairong zheng1,2
1Paul C. Lauterbur Research Centre for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 2University of Chinese Academy of Sciences, Beijing, China, 3Department of Magnetic Resonance Imaging, Fuwai Hospital and National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences, Peking Union Medical College, beijing, China
Cardiac magnetic resonance(CMR) is a highly reliable measurement to assess and predict cardiovascular events. The traditional regression model need a linear assumption but it can not be guaranteed. We use a ML method to predict cardiovascular events in HCM patients. According to our result, the C statistic for the ML model (0.804 [95% CI, 0.757-0.850]) was higher than Cox regression model (0.764, [95% CI, 0.671-0.857]). With the random sample, the ROC for the ML model(0.96 in the training set, 0.83 in the test set) was higher than the regression model(0.80 in the training set, 0.79 in the test set).
|
|||
2644.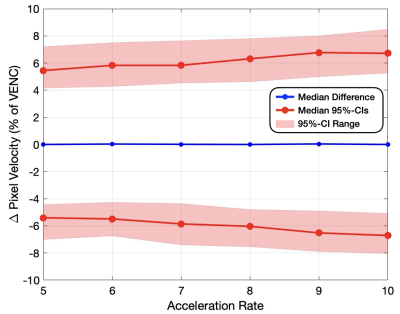 |
Cross Validation of a Deep Learning-Based ESPIRiT Reconstruction for Accelerated 2D Phase Contrast MRI
Jack R. Warren1, Matthew J. Middione2, Julio A. Oscanoa2,3, Christopher M. Sandino4, Shreyas S. Vasanawala2, and Daniel B. Ennis2,5
1Department of Computing + Mathematical Sciences, California Institute of Technology, Pasadena, CA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States, 3Department of Bioengineering, Stanford University, Stanford, CA, United States, 4Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 5Cardiovascular Institute, Stanford University, Stanford, CA, United States
Phase Contrast MRI (PC-MRI) measures the flow of blood. In order to obtain high-quality measurements, patients must hold their breath for ~20 seconds, which oftentimes can be difficult. Advances in deep learning (DL) have allowed for the reconstruction of highly undersampled MRI data. A 2D PC-MRI DL-ESPIRiT network was recently proposed to undersample the data acquisition by up to 8x without compromising clinically relevant measures of flow accuracy within ±5%. This work uses k-fold cross validation to evaluate the DL-ESPIRiT network on 2D PC-MRI data in terms of accuracy and variability for pixel velocity, peak velocity, and net flow.
|
|||
2645.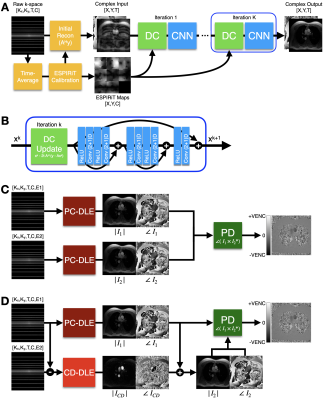 |
Deep Learning Based ESPIRiT Reconstruction for Highly Accelerated 2D Phase Contrast MRI
Julio A. Oscanoa1,2, Matthew J. Middione2, Christopher M. Sandino3, Shreyas S. Vasanawala2, and Daniel B. Ennis2,4
1Department of Bioengineering, Stanford University, Stanford, CA, United States, 2Department of Radiology, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 4Cardiovascular Institute, Stanford University, Stanford, CA, United States
We propose a novel Deep Learning (DL) based reconstruction framework for accelerated 2D Phase Contrast MRI (PC-MRI) datasets. We extend a previously developed DL method based on ESPIRiT reconstruction for cardiac cine and combine it with a direct Complex Difference estimation approach. We tested the DL methods using retrospectively undersampled 2D PC-MRI data and compared it with conventional Compressed Sensing (CS) reconstruction. Our method outperformed CS and enabled higher acceleration factors up to 8x while maintaining error metrics within a targeted accuracy of ±5%.
|
|||
2646.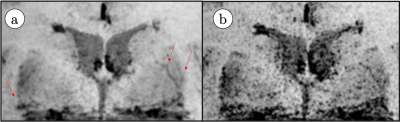 |
Exercise Effect in Human Brain Evaluated by 3D SWI Depiction of Lenticulostriate Artery with Denoising Deep Learning Reconstruction and 3D pCASL.
Vadim Malis1, Won Bae1, Asako Yamamoto1, Yoshimori Kassai2, Marin A McDonald1, and Mitsue Miyazaki1
1Radiology, UC San Diego, San Diego, CA, United States, 2Canon Medical, Tochigi, Japan
This study demonstrated that a mild exercise increases length and depiction of discernible lenticulostriate artery branches in 3D susceptibility weighted imaging, facilitated by applying denoising deep learning reconstruction (dDLR), and post-processed using vascular structure extraction algorithm (VSEA). Additionally, exercise increased intracranial perfusion, determined using 3D pCASL.
|
|||
2647.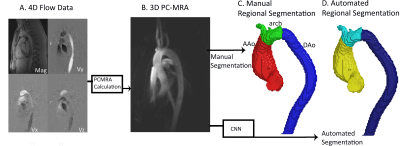 |
Deep Learning based Automatic Multi-Regional Segmentation of the Aorta form 4D Flow MRI
Haben Berhane1, Michael Scott1, Justin Baraboo1, Cynthia Rigsby2, Joshua Robinson2, Bradley Allen3, Chris Malaisrie3, Patrick McCarthy3, Ryan Avery3, and Michael Markl1
1Biomedical Engineering, Northwestern University, Chicago, IL, United States, 2Lurie Childrens Hospital of Chicago, Chicago, IL, United States, 3Northwestern Radiology, Evanston, IL, United States
While 4D flow MRI is capable of providing extensive hemodynamic quantifications, it requires cumbersome and time-consuming pre-processing. In order to accelerate the process, we developed and validated a multi-label convolutional neural network (CNN) for automatic aortic 3D segmentation and regional-labeling (ascending, AAo; arch; and descending aorta, DAo). Utilizing 320 4D flow MRI datasets, we used a 10-fold cross validation for training and testing the CNN. The Dice scores across each region were AAo: 0.95 [0.93-0.98], arch: 0.90 [0.89-0.95], and DAo: 0.95 [0.94-0.98]. Across all flow metrics, Bland-Altman comparisons showed moderate to excellent agreement between the manual and automated regional segmentations.
|
|||
2648.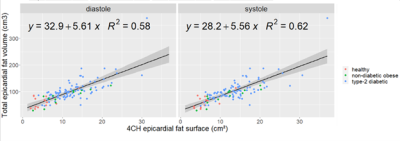 |
Deep-Learning epicardial fat quantification using 4-chambers Cardiac MRI segmentation, comparison with total epicardial fat volume
Pierre Daudé1, Patricia Ancel2, Sylviane Confort-gouny1, Anne Dutour2, Bénédicte Gaborit2, and Stanislas Rapacchi1
1Aix-Marseille Univ, CNRS, CRMBM, Marseille, France, 2APHM, Hôpital Universitaire Timone, Service d’Endocrinologie, Marseille, France
Evaluation of epicardial adipose tissue (EAT) burden holds potential as a biomarker for CHD diagnosis. EAT volume is challenging to assess using MRI due to its curved shape susceptible to partial volume effect. As a substitute, 4-chamber EAT surface can be reliably measured and has shown good correlation with EAT volume (r2=0.62). Two fully convolutional neural networks (FCN) were investigated for the segmentation of EAT surface on a database of 126 subjects. Promising results were obtained with DICE values of 0.71.
|
|||
2649.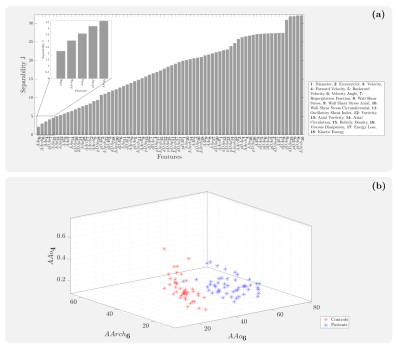 |
Identification of Hemodynamic Biomarkers for Bicuspid Aortic Valve Patients using Machine Learning
Pamela Franco1,2,3, Julio Sotelo1,3,4, Lydia Dux-Santoy5, Andrea Guala5, Aroa Ruiz-Muñoz5, Arturo Evangelista5, José Rodríguez-Palomares5, and Sergio Uribe1,3,6
1Biomedical Imaging Center, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 2Electrical Engineering Department, School of Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile, 3Millennium Nucleus for Cardiovascular Magnetic Resonance, Santiago, Chile, 4School of Biomedical Engineering, Universidad de Valparaíso, Valparaíso, Chile, 5Department of Cardiology, Hospital Universitari Vall d’Hebron, Vall d’Hebron Institut de Recerca (VHIR), Universitat Autònoma de Barcelona, Barcelona, Spain, 6Radiology Department, School of Medicine, Pontificia Universidad Católica de Chile, Santiago, Chile
The clinical significance and economic burden of bicuspid aortic valve (BAV) disease justify the need for improved clinical guidelines and more robust therapeutic modalities. Recent advances in medical imaging have demonstrated the existence of altered hemodynamics in these patients. To identify hemodynamic biomarkers for BAV patients, we present a machine learning method consisting of a feature selection mechanism to classify healthy volunteers and BAV patients accurately.
|
|||
2650.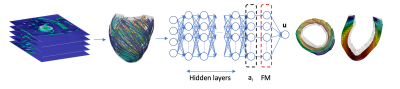 |
Fast personalization of cardiac mechanical models using parametric physics informed neural networks
Stefano Buoso1, Thomas Joyce1, and Sebastian Kozerke1
1ETH Zurich, Zurich, Switzerland
We propose a parametric physics-informed neural network that can be personalized to left-ventricular anatomies from cardiac MRI data. The model combines a left-ventricular anatomical shape model derived from cardiac MRI data, and a functional model derived from synthetic cardiac deformations. The network is trained with a label-free approach using a physics-based cost-function in less than 5 minutes on a single CPU. Network inputs are endocardium pressure and myocyte activation. A complete cardiac cycle can be simulated in less than a minute. This approach is 30 times faster than the corresponding finite element simulation even when including training time.
|
|||
2651.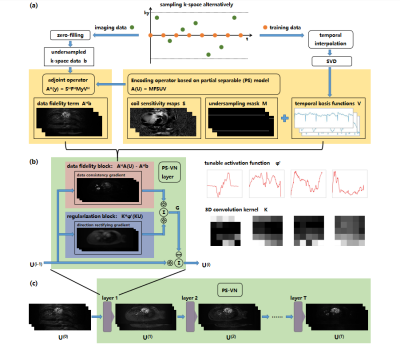 |
PS-VN: integrating deep learning into model-based algorithm for accelerated reconstruction of real-time cardiac MR imaging
Zhongsen Li1, Hanyu Wei1, Chuyu Liu1, Yichen Zheng1, Shuo Chen1, and Rui Li1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China
In this study, we combine classical “partial separable” model with deep-learning framework “variational network” for accelerated reconstruction of real-time cardiac MR imaging. The proposed PS-VN architecture achieves comparable reconstruction accuracy with baseline algorithm and reduce computational time to around 10 seconds for the reconstruction of over 4 thousand dynamic frames.
|
|||
2652.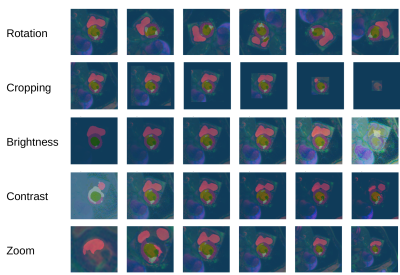 |
Sensitivity of a Deep Learning Model for Multi-Sequence Cardiac Pathology Segmentation to Input Data Transformations
Markus J Ankenbrand1, Liliya Shainberg1, Michael Hock1, David Lohr1, and Laura Maria Schreiber1
1Chair of Cellular and Molecular Imaging, Comprehensive Heart Failure Center (CHFC), University Hospital Würzburg, Würzburg, Germany
Deep learning-based segmentation models play an important role in cardiac magnetic resonance imaging. While their performance is good on the training and validation data the models themselves are hard to interpret. Sensitivity analysis helps to estimate the effect of different data characteristics on segmentation performance. We demonstrate that a published model exhibits higher sensitivity to basic transformations like rotation for pathology classes than for tissue classes in general.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.