Digital Posters
Emerging Applications of AI in Neuroimaging for CES II
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 6 | 19:00 - 20:00 |
 |
3501.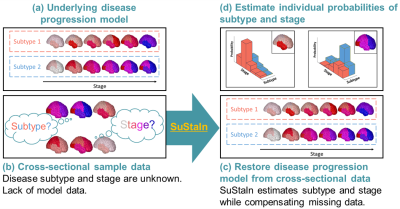 |
Temporal progression patterns of white-matter degeneration in CBS and PSP identified with Subtype & Stage Inference (SuStaIn)
Yuya Saito1, Peter A. Wijeratne2, Koji Kamagata1, Christina Andica1, Wataru Uchida1,3, Toshiaki Akashi1, Akihiko Wada1, Masaaki Hori4, and Shigeki Aoki1
1Department of Radiology, Juntendo University Graduate School of Medicine, Tokyo, Japan, 2Centre for Medical Image Computing, Department of Computer Science, University College London, London, United Kingdom, 3Department of Radiological Sciences, Graduate School of Human Health Sciences, Tokyo Metropolitan University, Tokyo, Japan, 4Department of Radiology, Toho University Omori Medical Center, Tokyo, Japan
Corticobasal syndrome (CBS) and progressive supranuclear palsy (PSP) are classic clinical syndromes derived from 4R tau pathology. Differential clinical diagnosis remains difficult due to neurodegenerative overlap. Most previous studies have assessed white-matter (WM) degeneration using cross-sectional data. This study applied Subtype & Stage Inference (SuStaIn), a novel unsupervised machine-learning technique for regional WM fractional anisotropy based on cross-sectional brain diffusion MRI to identify differences in temporal progression patterns of WM degeneration between CBS and PSP. Results suggested the utility of SuStaIn for identifying temporal WM degeneration patterns in and classifying patients with CBS and PSP.
|
||
3502.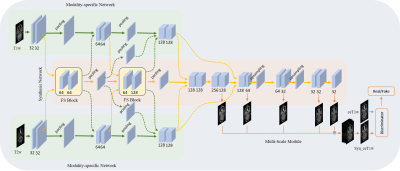 |
CE-Net: multi-inputs contrast enhancement network for nasopharyngeal carcinoma contrast enhanced T1-weighted MR synthesis
Wen Li1, Ge Ren1, Tian Li1, Haonan Xiao1, Francis Kar-ho Lee2, Kwok-hung Au2, and Jing Cai1
1Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, China, 2Department of Clinical Oncology, Queen Elizabeth Hospital, Hong Kong, China
To reduce the usage of gadolinium-based contrast agents (GBCAs), we proposed a deep learning based multi-inputs network (CE-Net) for contrast enhanced T1-weighted MR image synthesis based on pre-contrast T1-weighted and T2-weighted images in nasopharyngeal carcinoma (NPC) cases. When compared with multi-channel input methods, the proposed CE-Net has the ability to extract information from each input modalities separately. Supervision and multi-scale strategies are also applied in the proposed network. Quantitative and qualitative results show that our proposed CE-Net could achieve better performance when compared with the newly proposed Hi-Net and its extensions.
|
|||
3503. |
An Analysis of the Interpretability of Neural Networks trained on Magnetic Resonance Imaging for Stroke Outcome Prediction
Esra Zihni1, Bryony McGarry1,2, and John D. Kelleher1,3
1PRECISE4Q, Predictive Modelling in Stroke, Information Communications and Entertainment Institute, Technological University Dublin, Dublin, Ireland, 2School of Psychological Science, University of Bristol, Bristol, United Kingdom, 3ADAPT Research Centre, Technological University Dublin, Dublin, Ireland
Applying deep learning models to MRI scans of acute stroke patients to extract features that are indicative of short-term outcome could assist a clinician’s treatment decisions. Deep learning models are usually accurate but are not easily interpretable. Here, we trained a convolutional neural network on ADC maps from hyperacute ischaemic stroke patients for prediction of short-term functional outcome and used an interpretability technique to highlight regions in the ADC maps that were most important in the prediction of a bad outcome. Although highly accurate, the model’s predictions were not based on aspects of the ADC maps related to stroke pathophysiology.
|
|||
3504.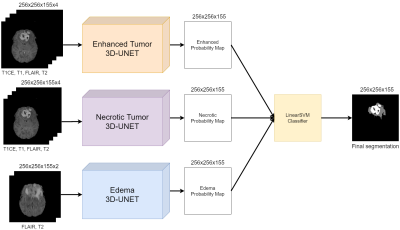 |
Volumetric assessment of patients with Glioblastoma by HUMBLe: Hierarchical 3D U-Net for MRI Brain Lesion segmentation
Yuval Buchsweiler1,2, Orna Aizenstein3,4, Felix Bokstein3,5,6, Idan Bressler1,2, Netanell Avisdris2,7, Deborah T. Blumenthal3,5, Dror Limon3,8, Dafna Ben Bashat2,3,6, and Moran Artzi2,3,6
1The Iby and Aladar Fleischman Faculty of Engineering, Tel Aviv University, Tel Aviv, Israel, 2Sagol Brain Institute, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 3Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 4Division of Radiology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 5Neuro-Oncology Service, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 6Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 7School of computer science and engineering, Hebrew University of Jerusalem, Israel, Jerusalem, Israel, 8Division of Oncology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel
Brain tumor segmentation is highly important for clinical management. We propose HUMBLe, a hierarchical 3D U-Net for MRI Brain Lesion segmentation architecture. HUMBLe breaks down the segmentation into its separate classes: enhancing tumor, edema, and necrotic classes, and uses a classifier to merge the different segmentation results into a final segmentation mask. Evaluation was performed on multi-parametric longitudinal local dataset, of patients with Glioblastoma. Segmentation results obtained by HUMBLe on our cohort improved DICE scores by 7%-16% for the different tumor components, compared to segmentation performed using 3D U-Net based architecture trained on BraTS2019 and our cohort.
|
|||
3505.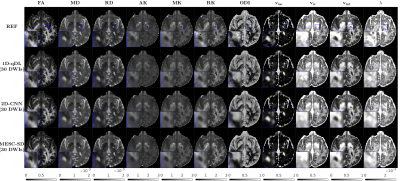 |
Rapid Estimation of Multiple Diffusion Maps from Undersampled Q-Space Data: A Comparison of Three Deep Learning Approaches
SeyyedKazem HashemizadehKolowri1,2, Rong-Rong Chen2, Ganesh Adluru1,3, and Edward V. R. DiBella1,2,3
1Radiology and Imaging Science, University of Utah, SALT LAKE CITY, UT, United States, 2Electrical and Computer Engineering, University of Utah, SALT LAKE CITY, UT, United States, 3Biomedical Engineering, University of Utah, SALT LAKE CITY, UT, United States
Advanced diffusion models enable characterization of tissue microstructure with higher specificity than conventional DTI. While beyond DTI diffusion imaging have been found valuable in many studies, their clinical availability have been hampered mainly due to their long scan times. Furthermore, each diffusion model can only extract a few relevant microstructural features. Therefore, using multiple models helps to better understand the brain microstructure, which requires multiple expensive model-fitting. In this study, we use different deep learning approaches to jointly estimate multiple advanced diffusion maps from highly undersampled q-space data, which can reduce both the scan and processing times significantly.
|
|||
3506.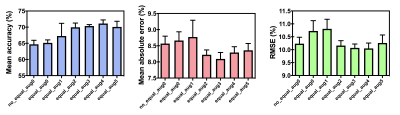 |
Deep learning prediction of retrieved stroke thrombus RBC content using quantitative, multiparametric MRI
Spencer D. Christiansen1,2, Junmin Liu2, Maria Bres Bullrich3, Manas Sharma4, Sachin K. Pandey4, Mel Boulton3, Luciano A. Sposato3, and Maria Drangova1,2
1Medical Biophyics, Western University, London, ON, Canada, 2Robarts Research Institute, London, ON, Canada, 3Clinical Neurological Sciences, London Health Sciences Centre, London, ON, Canada, 4Department of Medical Imaging, Western University, London, ON, Canada
Thrombus red blood cell (RBC) content has been associated with ischemic stroke etiology and responsiveness to recanalization therapies, yet currently can only be analyzed through retrospective histological analysis. We evaluated the ability of a convolutional neural network for predicting thrombus RBC content using multiparametric (R2*, QSM, late echo GRE) MR image slices of retrieved stroke thrombi ex vivo. The network predicted thrombus RBC content with an accuracy and mean absolute error of up to 71 and 8%, respectively, when data augmentation was applied. This technique holds potential for in vivo RBC content prediction and improving acute stroke care.
|
|||
3507.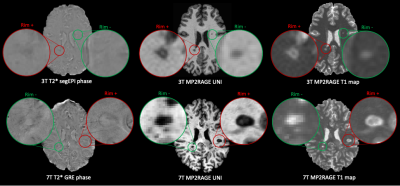 |
Automated assessment of paramagnetic rim lesions in multiple sclerosis patients with 3T and 7T MP2RAGE
Francesco La Rosa1,2,3, Germán Barquero1,2,3, Omar Al-Louzi4, Bénédicte Maréchal1,3,5, Tobias Kober1,3,5, Jean-Philippe Thiran1,3, Pascal Sati4,6, Daniel S. Reich4, Pietro Maggi7,8, Martina Absinta4,9, Meritxell Bach Cuadra1,2,3, and Cristina Granziera10,11,12
1Signal Processing Laboratory (LTS5), École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 2Medical Image Analysis Laboratory, Center for Biomedical Imaging (CIBM), University of Lausanne, Lausanne, Switzerland, 3Radiology Department, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 4Translational Neuroradiology Section, National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, MD, United States, 5Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 6Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, United States, 7Department of Neurology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 8Department of Neurology, Cliniques Universitaires Saint-Luc, Université Catholique de Louvain, Brussels, Belgium, 9Department of Neurology, Johns Hopkins University, Baltimore, MD, United States, 10Neurologic Clinic and Policlinic, Departments of Medicine, Clinical Research and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 11Translational Imaging in Neurology (ThINk) Basel, Department of Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 12Research Center for Clinical Neuroimmunology and Neuroscience (RC2NB) Basel, University Hospital Basel and University of Basel, Basel, Switzerland
Paramagnetic rim lesions (PRL) in multiple sclerosis are chronic inflammatory lesions depicted in susceptibility-based MRI where high PRL lesion burden has been associated with a more aggressive disease course. As their visual detection is subjective and time-consuming, a convolutional neural network (RimNet) has recently been developed and applied to 3T susceptibility contrast MR images. In this work, we evaluate a unimodal RimNet architecture based on either the MP2RAGE uniform contrast or the concurrently obtained T1 map at both 3T and 7T. Results show that prediction improves considerably at 7T, suggesting that 7T MP2RAGE might be helpful for automatically identifying PRL.
|
|||
3508.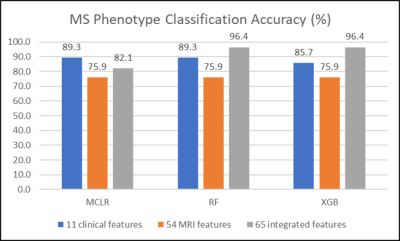 |
Performance evaluation of machine learning algorithms for multiple sclerosis phenotype classification using 7-Tesla MRI and clinical features
Seongjin Choi1 and Daniel M Harrison1,2
1Neurology, University of Maryland School of Medicine, Baltimore, MD, United States, 2Neurology, Johns Hopkins University School of Medicine, Baltimore, MD, United States
Three machine-learning algorithms were evaluated in the multiple sclerosis phenotype classification of a relatively small cohort. High accuracy of multiple-sclerosis phenotype classification was achievable by applying tree-based ensemble methods to integrated 7T MRI and clinical data features. Feature integration did not guarantee performance improvements in all machine learning algorithms evaluated. Features considered important may vary depending on the classification algorithm used.
|
|||
3509.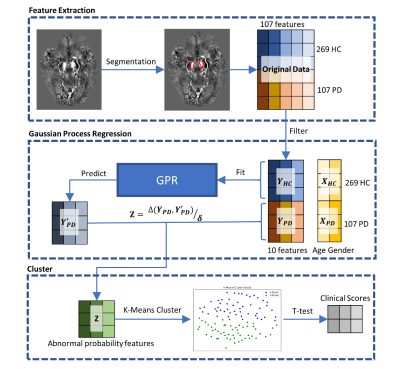 |
Parkinson’s Disease Subtype Identification Using Radiomics Based on Iron Deposition in Substantia Nigra
Zhijia Jin1, Ruiqi Yu2, Chenglong Wang2, Naying He1, Yan Li1, Zenghui Cheng1, Yida Wang2, Mark E. Haacke1,3,4,5, Guang Yang2, and Fuhua Yan1
1Department of Radiology, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China, 2Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, Shanghai, China, 3Magnetic Resonance Innovations, Inc., Bingham Farms, MI, United States, 4Department of Radiology, Wayne State University, Detroit, MI, United States, 5Department of Biomedical Engineering, Wayne State University, Detroit, MI, United States
A total of 104 Parkinson’s disease (PD) patients and 269 age- and sex-matched healthy controls (HCs) were scanned using 3D multi-echo gradient echo MTC sequence. In this work, a data-driven clustering approach based on iron deposition in the substantia nigra (SN) measured by quantitative susceptibility mapping (QSM) was performed to classify the PD patients into different subtypes. The clinical assessments were compared between subtype groups. Two subtypes were found by using this clustering approach. Furthermore, there are significant differences (p-values < 0.05) on MDS-UPDRS scores between these two subtype groups.
|
|||
3510.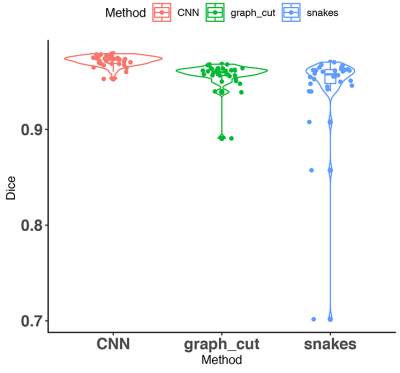 |
A comparison of three whole brain segmentation methods for in vivo manganese enhanced MRI in animal models of Alzheimer’s disease
Igor Varga1,2, Hae Sol Moon3, Adam Conrad4, Matthew Holbrook3, Andrei R Niculescu5, Abinaya Lakshmanan3, Robert J Anderson5, Christina L Williams6, Cristian T Badea3,5, Po-Wah So2, and Alexandra Badea3,5,7,8
1Department of Cybernetics, Czech Technical University, Prague, Czech Republic, 2Department of Neuroimaging, King's College London, London, United Kingdom, 3Biomedical Engineering, Duke University, Durham, NC, United States, 4Georgia Institute of Technology, Atlanta, GA, United States, 5Radiology, Duke University Medical Center, Durham, NC, United States, 6Psychology and Neuroscience, Duke University, Durham, NC, United States, 7Neurology, Duke University, Durham, NC, United States, 8Brain Imaging and Analysis, Duke University Medical School, Durham, NC, United States
Whole mouse brain segmentation is an essential prerequisite for multiple quantitative image analysis tasks and pipelines. In this work, we compare three methods for whole brain segmentation (skull stripping) relying on active contours, graph cuts and convolutional neural networks. We applied these methods on mouse brain manganese enhanced MR images acquired at 100 micrometre isotropic resolution, in vivo. All three methods achieved Dice coefficients larger than 94%, but convolutional neural networks achieved a small but significant improvement (0.97±0.01) over our active contours implementation (0.94±0.05) and the difference approached significance relative to graph cuts (0.96±0.01).
|
|||
3511.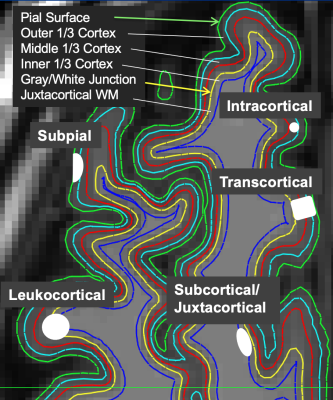 |
Novel machine learning method for clinically significant cortical lesion detection in multiple sclerosis
Eve L Kazarian1, Mariam S Aboian1, and John D Port2
1Department of Radiology and Biomedical Imaging, Yale University, New Haven, CT, United States, 2Department of Radiology, Mayo Clinic, Rochester, MN, United States
A novel tree-based machine learning model was applied to head MR imaging scans from multiple sclerosis (MS) patients in order to detect cortical gray matter lesions. Cortical lesions are strong indicators of MS disability yet are not easily visualized in clinical practice. The model was useful for accurately detecting cortical lesion presence (AUC of 0.78 ± 0.02).
|
|||
3512.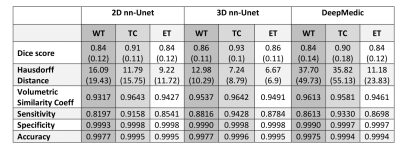 |
Multi-class Deep Learning Glioma Segmentation in Hospital Data with Missing Sequences
Jiaming Wu1, Hugh G. Pemberton1,2, Ivar Kommers3, Domenique M.J. Müller3, Sjoerd B. Vos1,2, Ferran Prados1,2, Yipeng Hu1, Pierre A. Robe4, Hilko Ardon5, Lorenzo Bello6, Marco Rossi6, Tommaso Sciortino6, Marco Conti Nibali 6,
Mitchel S. Berger7, Shawn L. Hervey-Jumper7, Wim Bouwknegt8, Wimar A. Van den Brink9, Julia Furtner10, Seunggu J. Han11, Albert J. S. Idema12, Barbara Kiesel13, Georg Widhalm13, Alfred Kloet14, Michiel Wagemakers15, Aeilko H. Zwinderman16, Sandro M. Krieg17,18,
Emmanuel Mandonnet19, Philip de Witt Hamer3, Roelant S. Eijgelaar3, and Frederik Barkhof1,20
1Centre for Medical Image Computing (CMIC), University College London, London, United Kingdom, 2Neuroradiological Academic Unit, UCL Queen Square Institute of Neurology, University College London, London, United Kingdom, 3Neurosurgical Center Amsterdam, Amsterdam UMC, Vrije Universiteit, Amsterdam, Netherlands, 4Department of Neurology & Neurosurgery, University Medical Center Utrecht, Utrecht, Netherlands, 5Department of Neurosurgery, St. Elisabeth Hospital, Tilburg, Netherlands, 6Neurosurgical Oncology Unit, Departments of Oncology and Hemato-Oncology, Università degli Studi di Milano, Humanitas Research Hospital, IRCCS, Milan, Italy, 7Department of Neurological Surgery, University of California, San Francisco, San Francisco, CA, United States, 8Department of Neurosurgery, Medical Center Slotervaart, Amsterdam, Netherlands, 9Department of Neurosurgery, Isala Hospital, Zwolle, Netherlands, 10Department of Biomedical Imaging and Image-Guided Therapy, Medical University Vienna, Vienna, Austria, 11Department of Neurological Surgery, Oregon Health and Science University, Portland, OR, United States, 12Department of Neurosurgery, Northwest Clinics, Alkmaar, Netherlands, 13Department of Neurosurgery, Medical University Vienna, Vienna, Austria, 14Department of Neurosurgery, Medical Center Haaglanden, The Hague, Netherlands, 15Department of Neurosurgery, University of Groningen, University Medical Center Groningen, Groningen, Netherlands, 16Department of Clinical Epidemiology and Biostatistics, Academic Medical Center, Amsterdam, Netherlands, 17TUM-Neuroimaging Center, Klinikum rechts der Isar, Technische Universität München, Munich, Germany, 18Department of Neurosurgery, Klinikum rechts der Isar, Technische Universität München, Munich, Germany, 19Department of Neurosurgery, Lariboisière Hospital, APHP, Paris, France, 20Radiology & Nuclear Medicine, VU University Medical Center, Amsterdam, Netherlands
Accurate segmentation and morphological assessment of glioma can guide treatment and support follow-up. The Brain Tumour Segmentation (BraTS) challenge has been instrumental in promoting research and comparing various automated segmentation algorithms. However, models in the challenge are trained and measured on a strictly curated and high-quality dataset, which is not representative of clinically acquired MRI data. Therefore, we have tested the generalisability of three network architectures from two of the top performing BraTS challenge models. We show the utility of these models in the presence of missing sequences and different scanners in multi-centre hospital data of varying quality.
|
|||
3513.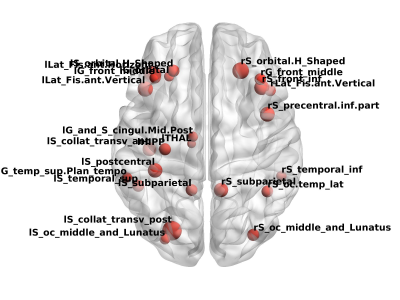 |
Aberrant white matter networks in methamphetamine-dependent patients and its application in support vector machine-based classification
Yadi Li1, Ping Cheng1, Pu-Yeh Wu2, Wenwen Shen3, Huifen Liu4, Jianbing Zhang4, Haibo Dong1, and Wenhua Zhou 3
1Department of Radiology, Ningbo Medical Treatment Center Lihuili Hospital, Ningbo University, Ningbo, China, 2GE Healthcare,Beijing,China, Beijing, China, 3Laboratory of Behavioral Neuroscience, Ningbo Addiction Research and Treatment Center, Ningbo, China, Ningbo, China, 4Ningbo Addiction Research and Treatment Center, Ningbo, China, Ningbo, China
This is a pilot study of the weighted white matter (WM) network in MA-dependent patients. By combining DTI-based probabilistic tractography and graph theory, the WM networks of MA-dependent patients presented small-worldness, and these networks tend to be random networks. The network metrics, that presented inter-group differences were used to construct a support vector machine, that achieved an excellent performance in discriminating MA-dependent patients from normal controls. Overall, the current study demonstrated that MA dependence is associated with abnormal network metrics, and these metrics can be promising features to train a classifier which need further verification with a larger sample size.
|
|||
3514.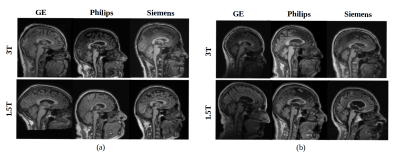 |
Machine Learning-based Analysis of Heterogeneous, Multi-center MR Datasets: Impact of Scan Variability
Mariana Bento1,2, Justin Park2,3, and Richard Frayne1
1Radiology and Clinical Neuroscience, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 2Calgary Image Processing and Analysis Centre, Foothills Medical Centre, Calgary, AB, Canada, 3Mechanical Engineering, University of Calgary, Calgary, AB, Canada
Multi-centre heterogeneous imaging datasets are frequently required to develop image-based computer-aided diagnosis and treatment monitoring tools. However, these datasets may present large underlying variability, potentially impacting the performance of the developed tools. Here, as a proof-of-concept, we propose a machine learning method to study scan variability related to the scanner vendor and magnetic field strength in brain MR images from two cohorts of healthy subjects. Our model has high accuracy rates (>92%), confirming the presence of scan variability in heterogeneous, multi-centre datasets. This model may be further incorporated into automated diagnostic tools, potentially allowing more reliable and robust results.
|
|||
3515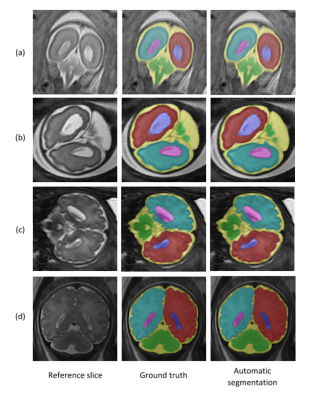 |
Automatic segmentation of fetal brain components from MRI using deep learning Video Permission Withheld
Ori Ben Zvi1,2, Netanell Avisdris1,3, Bossmat Yehuda1,2, Daphna Link Sourani1, Leo Joskowicz3, Elka Miller4, Liat Ben Sira2,5, and Dafna Ben Bashat1,2,5
1Sagol Brain Institute, Tel Aviv Sourasky Medical Center; Israel, Tel Aviv, Israel, 2Sagol School of Neuroscience, Tel Aviv University; Israel, Tel Aviv, Israel, 3School of Computer Science and Engineering, The Hebrew University of Jerusalem, Jerusalem, Israel, Jerusalem, Israel, 4Medical Imaging, Children’s Hospital of Eastern Ontario, University of Ottawa, Ottawa, Canada, Ottawa, ON, Canada, 5Sackler Faculty of Medicine, Tel Aviv University; Israel, Tel Aviv, Israel
Segmentation of the fetal brain into its components is important for quantitative assessment of fetal development. This study proposes a fully automatic method based on deep learning for fetal brain segmentation into six components, including a separation of right and left hemispheres. The method’s performance demonstrated high Dice scores for all brain components and robustness to different contrasts, scan resolutions, gestational age and fetal brain pathologies. Preliminary results demonstrated significant larger ventricle’s volumes and asymmetry in fetuses with ventriculomegaly compared to normal fetuses. The method is suggested to improve fetal assessment and assist radiologists in routine clinical practice.
|
|||
3516.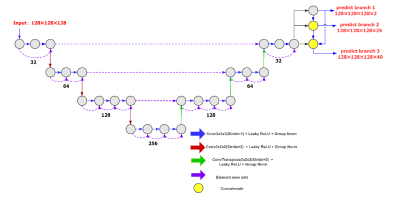 |
Task-aware 3D-Convolutional Neural Networks for Detailed Brain Parcellation
Junchuan Peng1, Yashi Nan1, Li Zhao2, Huanhui Xiao1, and Silun Wang1
1YIWEI Medical Technology Co., Ltd, Shenzhen, China, 2College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China
Morphological changes in neurodegenerative diseases can be detected with structural MR images, but it requires detailed brain parcellation. Therefore, a task-aware V-Net was proposed to segment the brain into 40 regions. Task-aware features were achieved by three cascading branches, including brain and non-brain regions, 25 regions with the bilateral regions grouped, and 40 regions, respectively. The proposed model was developed on 8938 subjects and validated using additional196 subjects. The proposed method outperformed the typical 3D U-Net and V-Net, and achieved state-of-the-art results on both datasets with a mean Dice score of 0.886 ± 0.029 and 0.874 ± 0.013, respectively.
|
|||
3517.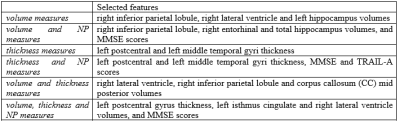 |
Brain atrophy and machine learning algorithms on the prediction of dementia development
Pedro Henrique Rodrigues da Silva1, Kaio Felippe Secchinato1, Julia Palaretti1, and Renata Ferranti Leoni1
1USP, Ribeirão Preto, Brazil
A challenging issue regarding the early diagnosis of the Alzheimer's disease (AD) is the selection of biomarkers. In this study, we aimed to classify cognitively normal elderly regarding the possibility to develop AD based on brain atrophy and neuropsychological scores, and using supervised machine learning algorithms. Our results suggest Naïve-Bayes (NB) classifiers with left postcentral and left middle temporal cortical thickness or right lateral ventricle, right inferior parietal and Corpus Callosum (CC) Mid Posterior volumes can be useful to identify in the early stage the subjects with higher risks to develop AD.
|
|||
3518. |
Clinical Application of Twelve-fold Accelerated Submillimeter Whole Brain 3D-T2 weighted Imaging with Deep Learning Reconstruction
Yasutaka Fushimi1, Satoshi Nakajima1, Akihiko Sakata1, Takuya Hinoda1, Sonoko Oshima1, Sayo Otani1, Krishna Pandu Wicaksono1, Hiroshi Tagawa1, Yang Wang1, Masahiro Nambu2, Rimika Imai2, Koji Fujimoto3, Hitomi Numamoto4, Kanae
Miyake4, Tsuneo Saga4, and Yuji Nakamoto1
1Kyoto University Hospital, Kyoto, Japan, 2MRI Systems Division, Canon Medical Systems Corporation, Otawara, Japan, 33. Department of Real World Data Research and Development, Kyoto University Graduate School of Medicine, Kyoto, Japan, 4Department of Advanced Medical Imaging Research, Kyoto University Graduate School of Medicine, Kyoto, Japan
Twelve-fold accelerated submillimeter 3D-T2 weighted imaging with DLR and imaging quality and sharpness of cranial nerves were examined.
|
|||
3519.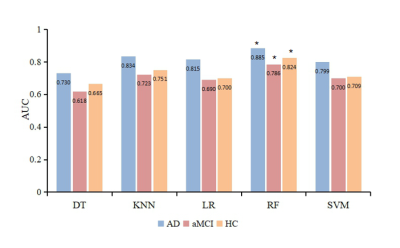 |
Machine Learning-based Features of DKI to Evaluate and Automate Alzheimer’s Disease and amnestic mild cognitive impairment Diagnoses
Yu Zhang1, Tongtong Li1, Xiuwei Fu2, Xianchang Zhang3, Yuan Luo4, and Hongyan Ni5
1First Central Clinical College, Tianjin Medical University, Tianjin, China, 2Tianjin Medical University General Hospital, Tianjin, China, 3MR Collaboration, Siemens Healthcare Ltd., Beijing, China, 4Department of Radiology, China-Japan Friendship Hospital, Beijing, China, 5Department of Radiology, Tianjin First Central Hospital, Tianjin, China
The early diagnoses of Alzheimer’s disease (AD) and amnestic mild cognitive impairment (aMCI) are crucial. This study aimed to acquire new imaging markers to assess the severity of AD and provide early diagnoses using machine learning algorithm. Diffusion kurtosis imaging (DKI) parameters were acquired on 58 AD patients, 64 aMCI patients, and 60 healthy volunteers. It’s found that radial diffusivity value of right uncinate fasciculus was the most important feature for assessing severity. The random forest classifier showed the highest diagnostic efficacy for AD. The RF classifier can provide an early diagnosis of disease based on the quantitative DKI features.
|
|||
3520.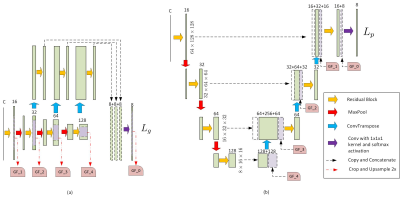 |
Brain Gray Matter Nuclei Segmentation on Quantitative Susceptibility Mapping using Convolutional Neural Network
Chao Chai1, Pengchong Qiao2, Bin Zhao2, Huiying Wang3, Guohua Liu2, Hong Wu2, E.Mark Haacke4, Wen Shen1, Xinchen Ye5, Zhiyang Liu2, and Shuang Xia1
1Department of Radiology, Tianjin First Central Hospital, Tianjin Medical Imaging Institute, School of Medicine, Nankai University, Tianjin, China, 2Tianjin Key Laboratory of Optoelectronic Sensor and Sensing Network Technology, College of Electronic Information and Optical Engineering, Nankai University, Tianjin, China, 3Department of Radiology, Tianjin First Central Hospital, School of Medicine, Nankai University, Tianjin, China, 4Department of Radiology, Wayne State University, Detroit, MI, United States, 5DUT-RU International School of Information Science and Engineering, Dalian University of Technology, Dalian, China
This study focused on developing an automatic gray matter nuclei segmentation method. A 3D convolutional neural network based method was proposed, which adopted patches with different resolutions as input for segmentation. Experimental results showed much higher segmentation accuracy over the atlas-based method and other deep-learning-based methods in terms of both the similarity and the surface distance metrics. The segmentation results of the proposed method is also evaluated in terms of measurement accuracy, where the proposed method achieves the highest consistency with the manual delineations.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.