Digital Posters
Explorations of AI in Neuroimaging
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 6 | 17:00 - 18:00 |
3265.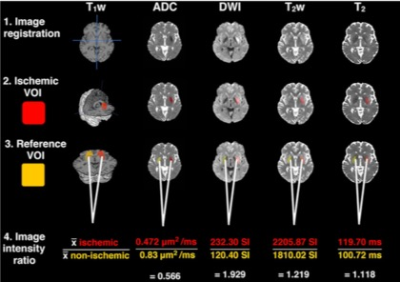 |
Stratifying ischaemic stroke patients across 3 treatment windows using T2 relaxation times, ordinal regression and cumulative probabilities
Bryony L. McGarry1,2, Elizabeth Hunter1, Robin A. Damion2, Michael J. Knight2, Philip L. Clatworthy3, George Harston4, Keith W. Muir5, Risto A. Kauppinen6, and John D. Kelleher1
1PRECISE4Q Predictive Modelling in Stroke, Information Communications and Entertainment Institute, Technological University Dublin, Dublin, Ireland, 2School of Psychological Science, University of Bristol, Bristol, United Kingdom, 3Stroke Neurology, North Bristol NHS Trust, Bristol, United Kingdom, 4Acute Stroke Programme, Radcliffe Department of Medicine, University of Oxford, Oxford, United Kingdom, 5Institue of Neuroscience and Psychology, University of Glasgow, Glasgow, United Kingdom, 6Faculty of Engineering, University of Bristol, Bristol, United Kingdom
Unknown onset time is a common contraindication for anti-thrombolytic treatment of ischaemic stroke.T2 relaxation-based signal changes within the lesion can identify patients within or beyond the 4.5-hour intravenous thrombolysis treatment-window. However, now that intra-arterial thrombolysis is recommended between 4.5 and 6 hours from symptom onset and mechanical thrombectomy is considered safe between 6 and 24 hours, there are three treatment-windows to consider. Here we show a cumulative ordinal regression model, incorporating the T2 relaxation time, predicts the probabilities of a patient being within one of the three treatment-windows and is more accurate than signal intensity changes from T2 weighted images.
|
|||
3266.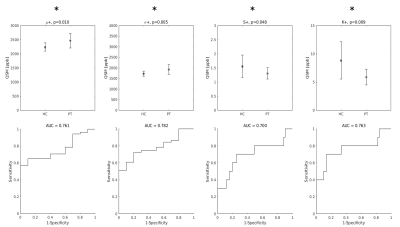 |
Distribution indices of QSM values in M1 enable to classify ALS patients and healthy controls
Mauro Costagli1,2, Graziella Donatelli3,4, Paolo Cecchi3,4, Gabriele Siciliano4,5, and Mirco Cosottini3,4,5
1University of Genova, Genova, Italy, 2IRCCS Stella Maris, Pisa, Italy, 3IMAGO 7 Research Foundation, Pisa, Italy, 4Azienda Ospedaliero Universitaria Pisana, Pisa, Italy, 5University of Pisa, Pisa, Italy
The Primary Motor Cortex (M1) in Amyotrophic Lateral Sclerosis (ALS) patients and healthy controls (HC) has a different appearance in Quantitative Susceptibility Mapping (QSM) images. The purpose of this study was to identify a set of distribution indices of QSM values in M1 that enable to better classify ALS patients and HC. Taken individually, the mean value, standard deviation, skewness and kurtosis of the QSM value distributions enabled to obtain diagnostic accuracies 0.43 < A < 0.78. When the distribution indices were jointly used in support vector machine (SVM) classifiers, it was possible to achieve a diagnostic accuracy of 0.90.
|
|||
3267.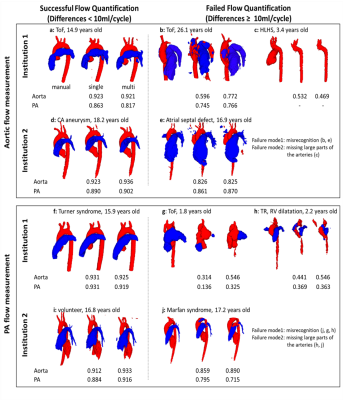 |
4D flow MRI hemodynamic quantification of pediatric patients with multi-site, multi-vender, and multi-channel machine learning segmentation
Takashi Fujiwara1, Haben Berhane2,3, Michael Baran Scott3, Zachary King2, Michal Schafer4, Brian Fonseca4, Joshua Robinson3, Cynthia Rigsby2,3, Lorna Browne4, Michael Markl3, and Alex Barker1,5
1Department of Radiology, Children's Hospital Colorado, University of Colorado Anschutz Medical Campus, Aurora, CO, United States, 2Lurie Children's Hospital of Chicago, Chicago, IL, United States, 3Northwestern University, Evanston, IL, United States, 4Children's Hospital Colorado, University of Colorado Anschutz Medical Campus, Aurora, CO, United States, 5Department of Bioengineering, University of Colorado Anschutz Medical Campus, Aurora, CO, United States
A convolutional neural network (CNN) is presented to quantify 4D flow MRI-based hemodynamics using automated segmentation of the proximal vasculature. The intent is to reduce time and user variability for cumbersome 4D flow MRI analyses; however, the pediatric setting is challenging given the complex arterial geometry often seen in congenital heart diseases. Multi-site and -vender datasets were used to train a CNN for 3D segmentation. Flow quantification was conducted with the automated segmentations to test if datasets from multiple institutions and vendors improves flow quantification. We found the multi-site approach improved flow measurements in the setting of complex disease.
|
|||
3268.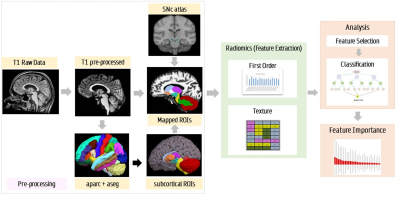 |
Delineating parkinsonian disorders using T1-weighted MRI based radiomics
Priyanka Tupe Waghmare1, Archith Rajan2, Shweta Prasad3, Jitender Saini4, Pramod Kumar Pal5, and Madhura Ingalhalikar6
1E &TC, Symbiosis Institute of Technology, Pune, India, 2Symbiosis Centre for Medical Image Analysis, Symbiosis Centre for Medical Image Analysis, Pune, India, 3Department of Clinical Neurosciences and Neurology, National Institute of Mental Health & Neurosciences, Bangalore, India, 4Department of Neuroimaging & Interventional Radiology, National Institute of Mental Health & Neurosciences, Bangalore, India, 5Department of Neurology, National Institute of Mental Health & Neurosciences, Bangalore, India, 6Symbiosis Center for Medical Image Analysis and Symbiosis Institute of Technology, Pune, India
Parkinson’s disease (PD), multiple system atrophy (MSA), and progressive supra-nuclear palsy (PSP) are neurodegenerative disorders which have parkinsonism as a core clinical feature. In the early stages PD and atypical parkinsonian syndrome (APS) (MSA and PSP) may often be indistinguishable and differential diagnosis is therefore crucial. Our work employs radiomics based features extracted from standard T1 weighted MRI images that are used in a machine learning framework to differentiate PD from APS. Results demonstrate a superior test accuracy of 92% that support our underlying hypothesis that radiomics on T1-weighted images can provide a discriminatory feature space between PD and APS.
|
|||
3269.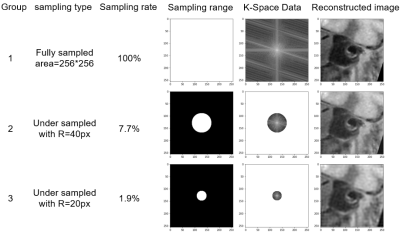 |
Automatic segmentation of arterial vessel wall on undersampled MR image using deep learning
Shuai Shen1,2,3,4, Xiong Yang5, Jin Fang6, Guihua Jiang6, Shuheng Zhang5, Yanqun Teng5, Xiaomin Ren5, Lele Zhao5, Jiayu Zhu5, Qiang He5, Hairong Zheng1,3,4, Xin Liu1,3,4, and Na Zhang1,3,4
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 2College of Software, Xinjiang University, Urumqi,, China, 3Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 4CAS key laboratory of health informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, shenzhen, China, 5Shanghai United Imaging Healthcare Co., Ltd., shanghai, China, 6Department of Radiology, Guangdong Second Provincial General Hospital, guangdong, China
A total of 124 patients were included in this study. We used U-net neural network architecture to segment the arterial vessel wall on original acquired MR vessel wall images and the corresponding images reconstructed from undersampled K-space data. The Dice coefficients based on the original K-space data, the K-space data with a sampling rate of 7.7%, and K-space data with a sampling rate of 1.9% were 88.66%, 88.19%, and 87.66%, respectively. The effectiveness of arterial vessel wall segmentation on undersampled images using U-net network was verified. The result demonstrated the potential to improve the acceleration performance of MR imaging.
|
|||
3270.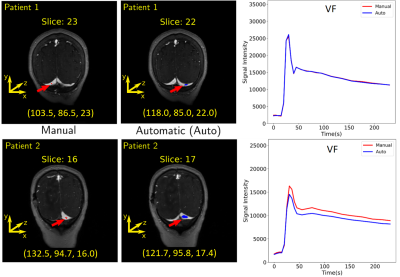 |
Automatic Vascular Function Estimation using Deep Learning for Dynamic Contrast-enhanced Magnetic Resonance Imaging
Wallace Souza Loos1,2, Roberto Souza2,3, Linda Andersen1,2, R. Marc Lebel2,4, and Richard Frayne1,2
1Radiology and Clinical Neuroscience, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 2Seaman Family MR Research Centre, Foothills Medical Centre, Calgary, AB, Canada, 3Electrical and Computer Engineering, Hotchkiss Brain Institute, University of Calgary, Calgary, AB, Canada, 4General Electric Healthcare, Calgary, AB, Canada
Dynamic contrast-enhanced magnetic resonance (DCE-MR) imaging is an important clinical tool for investigating the cerebrovascular microcirculatory systems including the blood-brain barrier through estimation of perfusion and permeability maps. However, a vascular function is needed to generate these maps. The estimation is usually performed manually, potentially leading to error, and is also time-consuming. In this work, we designed a deep learning model that leverages the temporal and spatial information from a time series of DCE-MR images to estimate the vascular function automatically. Our model was able to generalize well for unseen data and achieved good overall performance.
|
|||
3271.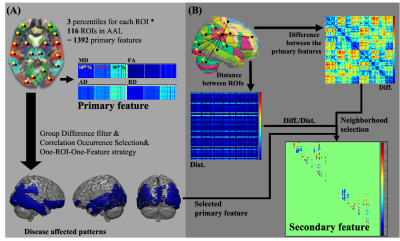 |
Pattern-based features extraction algorithm in the diagnosis of neurodegenerative diseases from diffusion MRI
Sung-han Lin1, Chih-Chien Tsai1, Yi-Chun Chen2,3, and Jiun-Jie Wang1
1Department of Medical Imaging and Radiological Sciences, Chang-Gung University, TaoYuan, Taiwan, 2Department of Neurology, Chang Gung Memorial Hospital Linkou Medical Center, TaoYuan, Taiwan, 3College of Medicine, Chang Gung University, TaoYuan, Taiwan
Improvement of diagnosis in patients with Mild Cognitive Impairment (MCI) would help in the early diagnosis of Alzheimer's Disease (AD), since MCI was known as the transition state from normal aging to AD. We developed a novel feature extraction algorithm, which was based on the disease clinical-pathological understanding and combined with the spatial information between disease affected pattern and its surrounding regions. This novel feature set demonstrated improved diagnostic performance, compared to the conventional feature set, especially for the MCI patients. In addition, the identified disease affected pattern corresponded with the postmortem pathology of amyloid deposition in AD patients.
|
|||
| 3272. | WITHDRAWN | |||
3273.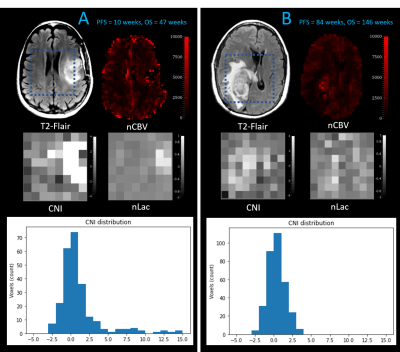 |
Early prediction of progression free survival and overall survival of patients with glioblastoma using machine learning and multiparametric MRI
Nate Tran1,2, Tracy Luks1, Devika Nair1, Angela Jakary1, Yan Li1, Janine Lupo1, Javier Villanueva-Meyer1, Nicholas Butowski3, Jennifer Clarke3, and Susan Chang3
1Department of Radiology & Biomedical Imaging, University of California, San Francisco, SAN FRANCISCO, CA, United States, 2UCSF/UC Berkeley Graduate Program in Bioengineering, SAN FRANCISCO, CA, United States, 3Department of Neurological Surgery, University of California, San Francisco, SAN FRANCISCO, CA, United States
This study evaluates the predictive power of multi-parametric MRI at pre-therapy and mid-RT time points in predicting progression-free and overall survival of patients with glioblastoma (GBM). We trained and tested random forest models using metabolic, perfusion, and diffusion images at both preRT and midRT scans, and found that not confining these metrics to the anatomical lesion boundaries improved outcome prediction. The CEL volume mid-RT and type of treatment were among the most important features in predicting PFS, while the T2L volume and metabolic metrics at pre-RT were more relevant for OS prediction.
|
|||
3274.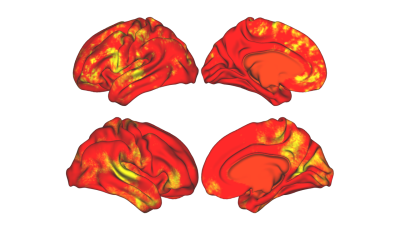 |
Exploring Brain Regions Involved in Working Memory using Interpretable Deep Learning
Mario Serrano-Sosa1, Jared Van Snellenberg2, and Chuan Huang2,3
1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Psychiatry, Renaissance School of Medicine at Stony Brook University, Stony Brook, NY, United States, 3Radiology, Renaissance School of Medicine at Stony Brook University, Stony Brook, NY, United States
Although deep learning algorithms are a novel method for neuroimage analysis, at times they are used as a “black box” for classification task. Therefore, it is crucial to develop methods to comprehend the abstract features used for prediction. We have developed an interpretable deep learning algorithm to predict working memory scores from fMRI data; wherein prediction performance was compared to Kernel Ridge Regression, a traditional machine learning approach. Across all metrics of evaluation, our method outperformed KRR. Moreover, our method was able to create averaged saliency maps highlighting regions most predictive of working memory scores.
|
|||
3275.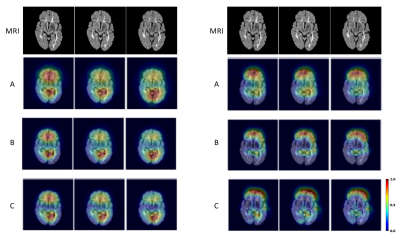 |
Multi-layer backpropagation of classification information with Grad-CAM to enhance the interpretation of deep learning models
Daphne Hong1 and Yunyan Zhang1
1University of Calgary, Calgary, AB, Canada
Deep learning is becoming increasingly important in medical imaging analysis, but the ability to interpret deep learning models still lag behind. Here, based on a promising method, gradient-weighted class activation mapping (Grad-CAM), we developed new approaches to interpret arbitrary layers of a convolutional neural network (CNN). Further, using two common CNN models trained to classify brain MRI scans into 3 types, we demonstrated the promise of our new strategy. Characterizing features at low and high levels of a CNN may provide new biomarkers and new insight into disease mechanisms, deserving further validation.
|
|||
3276. |
Improved Outcome prediction in mild Traumatic Brain Injury using Latent Feature Extraction from Volumetric MRI
Sanjay Purushotham1, Ashwathy Samivel Sureshkumar1, Li Jiang2, Shiyu Tang2, Steven Roys2, Chandler Sours Rhodes2,3, Rao P. Gullapalli2, and Jiachen Zhuo2
1Department of Information System, University of Maryland, Baltimore County, Baltimore, MD, United States, 2Diagnostic Radiology and Nuclear Medicine, University of Maryland School of Medicine, Baltimore, MD, United States, 3National Intrepid Center of Excellence, Walter Reed National Military Medical Center, Bethesda, MD, United States
Mild traumatic brain injury (mTBI) patients account for more than 70% of all TBI, with a subset experiencing persistent post concussive symptoms more than 3 months post injury. Patients clinical presentation and conventional imaging findings acutely post injury often lack the ability to predict chronic outcome. In this study, we present a novel method for latent feature extraction from volumetric MRI. We show that with machine learning methods, these acute latent volumetric MRI features are able to improve our symptom prediction in mTBI patients at 18 months post injury.
|
|||
3277.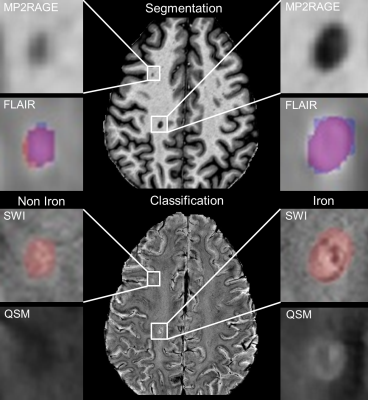 |
Prediction of iron rim lesions in multiple sclerosis using convolutional neural networks and multi-contrast 7T MRI data
René Schranzer1,2, Steffen Bollmann3, Simon Hametner2, Christian Menard1, Siegfried Trattnig4, Fritz Leutmezer2, Paulus Stefan Rommer2, Thomas Berger2, Assunta Dal-Bianco2, and Günther Grabner1,2,4
1Department of Medical Engineering, Carinthia University of Applied Sciences, Klagenfurt, Austria, 2Department of Neurology, Medical University of Vienna, Vienna, Austria, 3School of Information Technology and Electrical Engineering, The University of Queensland, Brisbane, Australia, 4Department of Biomedical Imaging and Image-guided Therapy, High Field Magnetic Resonance Centre, Vienna, Austria
In multiple sclerosis (MS) the presence of paramagnetic iron rim lesions has been shown to be indicative for progression with a more severe disease course. Our goal was to develop a pipeline based on neural networks to automatically detect, segment and classify lesions as either non-iron or iron loaded using multi-contrast 7T MRI data. A patch-based approach with two modified u-net architectures was used for segmentation and classification. Automatic, high quality lesion segmentation and their classification based on the presence or absence of iron-rims is enabled using convolutional neural networks.
|
|||
3278.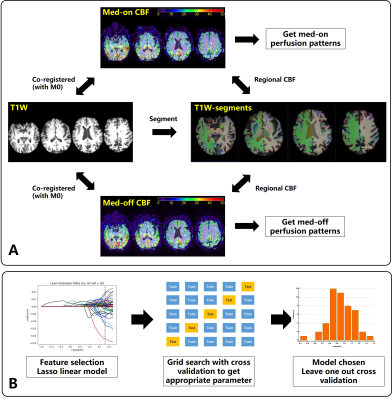 |
MRI-ASL Perfusion patterns may predict deep brain stimulation outcome in de novo Parkinson’s Disease
Hanyu Wei1, Le He1, Rongsong Zhou2, Shuo Chen1, Miaoqi Zhang1, Wenwen Chen1, Xuesong Li3, Yu Ma2, and Rui Li1
1Center for biomedical imaging research, Tsinghua University, Beijing, China, 2Department of Neurosurgery, Tsinghua University Yuquan Hospital, Beijing, China, 3School of Computer Science and Technology, Beijing Institute of Technology, Beijing, China
Deep brain stimulation(DBS) is routine treatment in Parkinson’s disease(PD) but its outcome varies among patients. It’s of benefit to investigate predictive value of pre-operative MR perfusion patterns in DBS outcome. In this study, regional cerebral blood flow(CBF) were measured by ASL both medication “on” and “off” before DBS surgery. Machine learning models were applied to select features and to predict the DBS outcome. Pre-operative MR perfusion patterns shows valuable to predict the DBS outcome in de novo PD patients. The Ridge regression performed best with good correlation(r=0.87,p<0.001) and good agreement with the measured DBS outcome at error level of 9.0%.
|
|||
3279.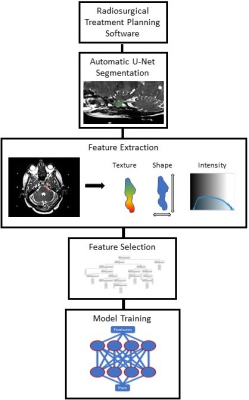 |
Using MRI and Radiomics to Predict Pain in a Cohort of Trigeminal Neuralgia Patients Treated With Radiosurgery
Kellen Mulford1, Sean Moen2, Andrew W. Grande2, Donald R. Nixdorf3, and Pierre-Francois Van de Moortele1
1Center for Magnetic Resonance Imaging, University of Minnesota, Minneapolis, MN, United States, 2Department of Neurosurgery, University of Minnesota, Minneapolis, MN, United States, 3Department of Diagnostic and Biological Science, University of Minnesota, Minneapolis, MN, United States
Due to a lack of objective markers, trigeminal neuralgia is difficult to diagnose and classify. This difficulty results in frequent pain recurrences following medication and invasive therapies. In this work, we show that a radiomics based model with features extracted from MRI images can be used to identify painful nerves. Using a cohort of trigeminal neuralgia patients, our predictive model achieves an accuracy of 78% and an AUC of 0.84 in distinguishing between nerves affected and nonaffected by trigeminal neuralgia.
|
|||
3280.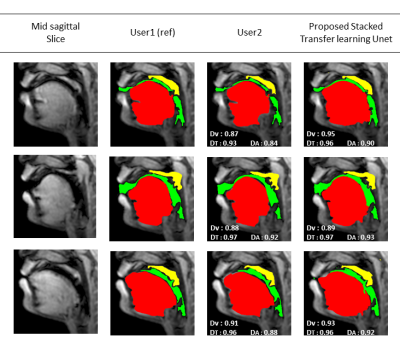 |
Stacked hybrid learning U-NET for segmentation of multiple articulators in speech MRI
SUBIN ERATTAKULANGARA1, KARTHIKA KELAT2, JUNJIE LIU3, and SAJAN GOUD LINGALA1,4
1Roy J Carver Department of Biomedical Engineering, University of Iowa, Iowa City, IA, United States, 2Government Engineering College Kozhikode, Kozhikode, India, 3Department of Neurology, University of Iowa, Iowa City, IA, United States, 4Department of Radiology, University of Iowa, Iowa City, IA, United States
We propose a stacked U-NET architecture to automatically segment the tongue, velum, and airway in speech MRI based on hybrid learning. Three separate U-nets are trained to learn the mapping between the input image and their specific articulator. The two U-NETs to segment the velum, and tongue are based on transfer learning, where we leverage open-source brain MRI segmentation. The third U-NET for airway segmentation is based on classical training methods. We demonstrate the utility of our approach by comparing against manual segmentations.
|
|||
3281.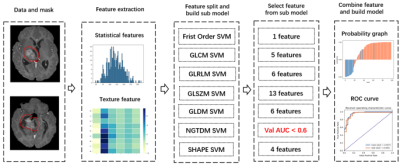 |
Use scout models for effective dimension reduction and feature selection in radiomics study
Yibo Dan1, Hongyue Tao2, Yida Wang1, Chengxiu Zhang1, Chenglong Wang1, Shuang Chen2, and Guang Yang1
1Shanghai Key Laboratory of Magnetic Resonance, East China Normal University, shanghai, China, 2Department of Radiology, Huashan Hospital, Fudan University, shanghai, China
Radiomics has been used widely in image-based diagnosis and prognosis. Since radiomics studies often involve a small number of samples, effective dimension reduction and feature selection are crucial to the successful modeling. In this study, we proposed a heuristic method for effective dimension reduction and feature selection, which built a scout model for each category of features to select features from the category for the final model building. The approach was applied to the modeling with two different datasets, including the BraTS 2019 open data, and achieved results better than those of traditional methods on both datasets.
|
|||
3282.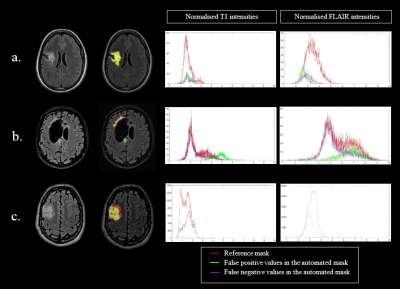 |
EVALUATION OF A CONVOLUTIONAL NEURAL NETWORK FOR AUTOMATED SEGMENTATION OF LOW-GRADE GLIOMAS
Margaux Verdier1,2, Justine Belko1, Jeremy Deverdun1, Nicolas Menjot de Champfleur1,3, Thomas Troalen2, Bénédicte Maréchal4,5,6, Emmanuelle Le Bars1, and Till Huelnhagen4,5,6
1I2FH , Neuroradiology, CHU Montpellier, Montpellier University, France, Montpellier, France, 2Siemens Healthcare, Saint Denis, France, 3Laboratoire Charles Coulomb, University of Montpellier, France, Montpellier, France, 4Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 5LTS5, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 6Radiology Department, Lausanne University Hospital and University of Lausanne, Switzerland, Lausanne, Switzerland
Tumor growth exceeding 8mm/year is the main indication for surgical intervention in low-grade gliomas (LGG). As manual growth assessment is very time-consuming, automated segmentation is desirable. We trained a Convolutional Neural Network (CNN) to segment LGG on 277 MRI-exams (T1+T2-FLAIR) and tested its performance on 9 unknown exams. The mean Dice Similarity Coefficient for automated segmentation was 0.72. The algorithm correctly segmented low T1 and high FLAIR values but tended to underestimate heterogeneous gliomas. Results were independent of cavity or tumor volume. Automated segmentation using CNNs seems promising for clinical practice. Performance might be improved using 3D FLAIR sequences.
|
|||
3283.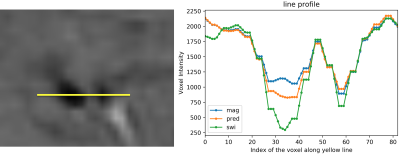 |
IMPROVING THE CONTRAST OF CEREBRAL MICROBLEEDS ON T2*-WEIGHTED IMAGES USING DEEP LEARNING
Ozan Genc1, Sivakami Avadiappan1, Yicheng Chen2, Christopher Hess1, and Janine M. Lupo1
1Department of Radiology and Biomedical Imaging, University of California San Francisco, San Francisco, CA, United States, 2Facebook Inc., Mountain View, CA, United States
This study trains a deep convolutional neural network to learn the SWI contrast from T2*-weighted magnitude images using a LSGAN deep learning model and assesses the performance of the resulting network on CMB detection. Our predicted SWI images were able to improve CMB contrast over T2* magnitude images and minimize residual artifacts from standard SWI processing.
|
|||
3284.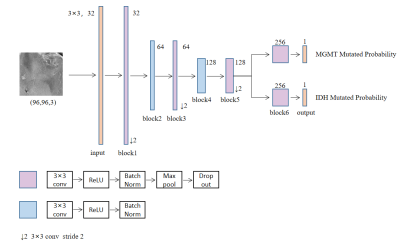 |
Automatic Prediction of MGMT and IDH Genotype for Gliomas from MR Images via Multi-task Deep Learning Network
Xiaoyuan Hou1,2, Hui Zhang1,2, Yan Tan3, Zhenchao Tang1,2, Hui Zhang3, and Jie Tian1,2
1Beijing Advanced Innovation Center for Big Data-Based Precision Medicine(BDBPM) ,Beihang University,100083, Beijing, China, 2Key Laboratory of Molecular Imaging, Institute of Automation, Chinese Academy of Sciences,100190, Beijing, China, 3Department of Radiology, First Clinical Medical College, Shanxi Medical University,030001, Taiyuan, China
In order to preoperatively predict the multiple genotype mutation for gliomas, we proposed an end-to-end multi-task deep learning model based on MR images analysis for simultaneously predicting IDH and MGMT mutation. Best-performed model was obtained by changing the number of sharing layers in the network, achieving accuracy of 79.78% for MGMT, 78.88% for IDH in the test dataset. Our results indicated that multi-task deep learning model provided a potential solution for simultaneously prediction of multiple genotype in gliomas.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.