Oral
Machine Learning Applications in CV Imaging
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 2 | 12:00 - 14:00 | Moderators: Aurelien Bustin & Kelvin Chow |
 |
0447.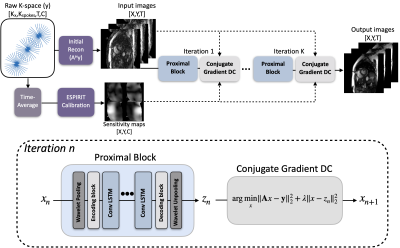 |
Improving deep unrolled neural networks for radial cine cardiac image reconstruction using memory-efficient training, Conv-LSTM based network
Kanghyun Ryu1, Christopher M. Sandino1, Zhitao Li1, Xucheng Zhu2, Andrew Coristine3, Martin Janich4, and Shreyas S. Vasanawala1
1Stanford University, Stanford, CA, United States, 2GE Healthcare, Menlo Park, CA, United States, 3GE Healthcare, Montreal, QC, Canada, 4GE Healthcare, Munich, Germany
Recently, unrolled neural networks (UNNs) have been shown to improve reconstruction over conventional Parallel Imaging Compressed Sensing (PI-CS) methods for dynamic MR image reconstruction. In this work we propose two methods to improve UNN for Non-Cartesian cardiac cine image reconstruction, namely memory efficient training and Convolutional LSTM based network architecture.The proposed method can significantly improve conventional UNN with higher image quality.
|
|
 |
0448.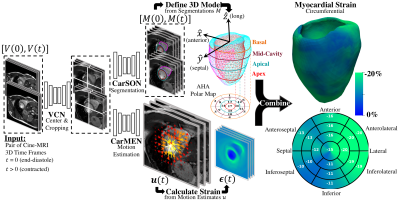 |
Development, Validation, and Application of an Automated Deep Learning Workflow for Strain Analysis based on cine-MRI
Manuel A. Morales1,2, Maaike van den Boomen2,3,4, Christopher Nguyen2,4, Jayashree Kalpathy-Cramer2, Bruce R. Rosen1,2, Collin Stultz 1,5,6, David Izquierdo-Garcia1,2, and Ciprian Catana2
1Health Sciences & Technology, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Radiology, Athinoula A. Martinos Center for Biomedical Imaging, MGH, HMS, Charlestown, MA, United States, 3Radiology, University Medical Center Groningen, Groningen, Netherlands, 4Cardiovascular Research Center, MGH, HMS, Charlestown, MA, United States, 5Cardiology, Massachusetts General Hospital, Boston, MA, United States, 6Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States
Myocardial strain analysis from cinematic magnetic resonance imaging (cine-MRI) data is a promising technique for earlier detection of subclinical dysfunction prior to reduction in left-ventricular ejection fraction (LVEF), but sources of discrepancies including user-related variations have limited its wide clinical adoption. Using healthy and cardiovascular disease (CVD) subjects (n=150) we developed a fast, user-independent deep-learning-based workflow for strain analysis from cine-MRI data. Relative to a reference tagging-MRI method, there was no significant difference in end-systolic global strain based on subject-paired cine-MRI data from 15 heathy subjects. Applications in CVD subjects without reduced LVEF showed both global and asymmetric strain abnormalities.
|
|
0449.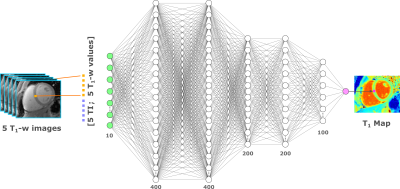 |
MyoMapNet: A Deep Neural Network for Accelerating the Modified Look-Locker Inversion Recovery Myocardial T1 Mapping to 5 Heart Beats
Hossam El-Rewaidy1,2, Rui Guo1, and Reza Nezafat1
1Medicine, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, United States, 2Graduate School of Bioengineering, Department of Computer Science, Technical University of Munich, Munich, Germany
In this work, we developed and evaluated a rapid (4-5 heartbeats) myocardial T1 mapping approach by estimating voxel-wise T1 values from one look-locker (LL) experiment of MOLLI sequence using a fully-connected neural network (MyoMapNet). MyoMapNet consists of 5 hidden layers that map the input 4-5 T1-weighted samplings and their inversion times into T1 values. MyoMapNet was trained and evaluated on a large dataset of native MOLLI-5(3)3 T1 in 717 subjects and post-contrast MOLLI-4(1)3(1)2 in 535 subjects. MyoMapNet showed similar T1 estimations to MOLLI-5(3)3 and MOLLI-4(1)3(1)2 T1 (mean difference=1±17ms, and -3±18ms, respectively, p-value >0.1 for both).
|
||
 |
0450.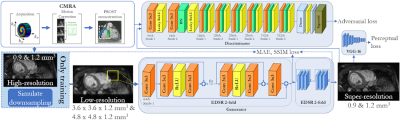 |
Deep-learning based super-resolution reconstruction for 3D isotropic coronary MR angiography in a one-minute scan
Thomas Küstner1,2, Alina Psenicny1, Camila Munoz1, Niccolo Fuin3, Aurelien Bustin4, Haikun Qi1, Radhouene Neji1,5, Karl P Kunze1,5, Reza Hajhosseiny1, Claudia Prieto1, and René M Botnar1
1School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 2Department of Radiology, Medical Image and Data Analysis (MIDAS), University Hospital of Tübingen, Tübingen, Germany, 3Ixico, London, United Kingdom, 4IHU LIRYC, Electrophysiology and Heart Modeling Institute, Université de Bordeaux, INSERM, Centre de recherche Cardio-Thoracuique de Bordeaux, Bordeaux, France, 5MR Research Collaborations, Siemens Healthcare Limited, Frimley, United Kingdom
3D whole‐heart coronary MR angiography (CMRA) has shown significant potential for the diagnosis of coronary artery disease. Undersampled motion corrected reconstruction approaches have enabled free-breathing isotropic 3D CMRA in ~5-10min scan time. However, spatial resolution is still limited compared to coronary CT angiography and scan time remains relatively long. In this work, we propose a deep-learning based super-resolution (SR) framework, combined with non-rigid respiratory motion compensation (SR-CMRA), to shorten the acquisition time to <1min. A 16-fold increase in spatial resolution is achieved by reconstructing a high-resolution CMRA (1.2mm3) from a low-resolution acquisition (1.2x4.8x4.8mm3, 50s scan).
|
|
 |
0451.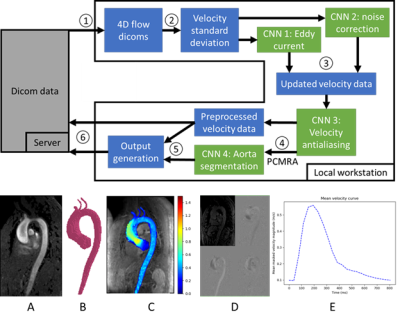 |
Fully automated aortic 4D flow MRI large-cohort analysis using deep learning
Michael B Scott1, Haben Berhane1, Justin Baraboo1, Cynthia K Rigsby2, Joshua D Robinson2, Patrick M McCarthy1, S Chris Malaisrie1, Ryan J Avery1, Bradley D Allen1, Alexander Barker3, and Michael Markl1
1Northwestern University, Chicago, IL, United States, 2Lurie Children's Hospital of Chicago, Chicago, IL, United States, 3University of Colorado, Anschutz Medical Campus, Aurora, CO, United States
A fully automated pipeline using four convolutional neural networks was designed to perform analysis of aortic 4D flow MRI, including preprocessing (eddy current correction, noise masking, and antialiasing), 3D segmentation of the aorta, quantification of mean flow-time curves and peak velocities. The analysis pipeline was run on a total of 2084 4D flow MRI studies and compared against manual analysis in a subset of 69 studies. Median segmentation Dice score for the ascending aorta was 0.93 [0.90 – 0.95]. Pipeline-based quantification of ascending aortic peak velocities demonstrated bias of -0.05 m/s versus manual analysis [LOA: -0.26 to 0.15 m/s].
|
|
0452.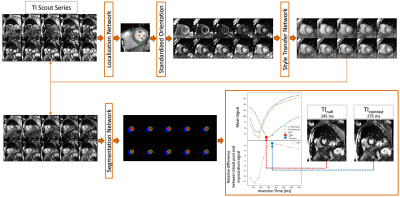 |
Validation of a Deep Learning based Automated Myocardial Inversion Time Selection for Late Gadolinium Enhancement Imaging in a Prospective Study
Seung Su Yoon1,2, Michaela Schmidt2, Manuela Rick2, Teodora Chitiboi3, Puneet Sharma3, Tilman Emrich4,5, Christoph Tilmanns6, Ralph Waßmuth6, Jens Wetzl2, and Andreas Maier1
1Pattern Recognition Lab, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Magnetic Resonance, Siemens Healthcare GmbH, Erlangen, Germany, 3Siemens Medical Solutions USA, Inc., Princeton, NJ, United States, 4Department of Radiology, University Medical Center, Johannes Gutenberg-University Mainz, Mainz, Germany, 5Division of Cardiovascular Imaging, Department of Radiology and Radiological Science, Medical University of South Carolina, Charleston, SC, United States, 6Diagnostikum Berlin, Berlin, Germany
In cardiac MRI using the Late Gadolinium Enhancement technique, inversion recovery sequences are acquired for the correct myocardial nulling for optimal image contrast. In clinical practice, the selection of the proper inversion time to null healthy myocardium is manually performed by visual inspection. To standardize the process, we propose an automated deep-learning-based system which selects the “null inversion time” where the myocardium signal is darkest, and “contrast inversion time” where the contrast between the myocardium and blood pool is highest. We validated the system on a prospective study on different scanners. The system achieved high accuracy in observers’ annotation range.
|
||
0453.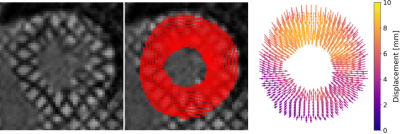 |
Voxel-wise Tracking of Grid Tagged Cardiac Images using a Neural Network Trained with Synthetic Data
Michael Loecher1,2, Luigi E Perotti3, and Daniel B Ennis1,2,4,5
1Radiology, Stanford University, Stanford, CA, United States, 2Radiology, Veterans Affairs Health Care System, Palo Alto, CA, United States, 3Mechanical and Aerospace Engineering, University of Central Florida, Orlando, FL, United States, 4Maternal & Child Health Research Institute, Stanford University, Stanford, CA, United States, 5Cardiovascular Institute, Stanford University, Stanford, CA, United States
This work introduces a neural network for tracking myocardial motion in cine grid tagged MR images on a voxel-wise basis. This is achieved with the use a synthetic training dataset that includes comprehensive motion patterns. Synthetic training allows for a known ground truth motion to be included in training. The network was tested against a previous network that tracked only tag line intersections. Displacements and strain maps were generated and compared. The voxel tracking network shows qualitatively better spatial localization of strain, and better radial strain values compared to tracking only tag lines.
|
||
0454.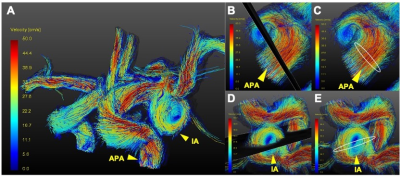 |
Prediction of aneurysm stability using a machine learning model based on 4D-Flow MRI and Black Blood MRI
Miaoqi Zhang1, Mingzhu Fu1, Hanyu Wei1, Shuo Chen1, and Rui Li1
1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China
The rupture of intracranial aneurysm (IA) is the most common cause of subarachnoid hemorrhage (SAH), resulting in patient death and disability. Recently, the morphology of the aneurysm, wall condition as well as hemodynamic factors were found to have kind of relationship with the stability of the aneurysm. In this project, we extracted clinical characteristics, morphology parameters, wall condition and hemodynamic parameters together to predict aneurysm stability using a machine learning model based on 4D-Flow MRI and black blood MRI. Among the two models, the Support Vector Machines model performed well, and the multi-parameter prediction result was greater than 95%.
|
||
0455.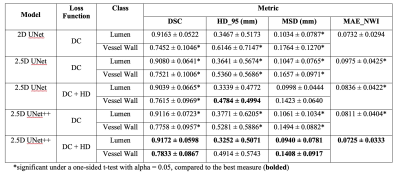 |
Intracranial Vessel Wall Segmentation with 2.5D UNet++ Deep Learning Network
Hanyue Zhou1, Jiayu Xiao2, Debiao Li1,2, Dan Ruan1,3, and Zhaoyang Fan2,4,5
1Bioengineering, University of California, Los Angeles, Los Angeles, CA, United States, 2Cedars-Sinai Medical Center, Los Angeles, CA, United States, 3Radiation Oncology, University of California, Los Angeles, Los Angeles, CA, United States, 4Radiology, University of Southern California, Los Angeles, CA, United States, 5Radiation Oncology, University of Southern California, Los Angeles, CA, United States
Intracranial vessel wall segmentation is an essential step for the intracranial atherosclerosis quantification. We have developed an automated intracranial vessel wall segmentation method based on deep learning that utilized a 2.5D UNet++ network structure with a loss function consists of both soft Dice coefficient loss and the approximated Hausdorff distance loss. We show that we have achieved significant improvements over our previous segmentation model based on a 2D UNet structure across various quantitative measures, as well as a better visual resemblance to the ground truth segmentation.
|
||
0456.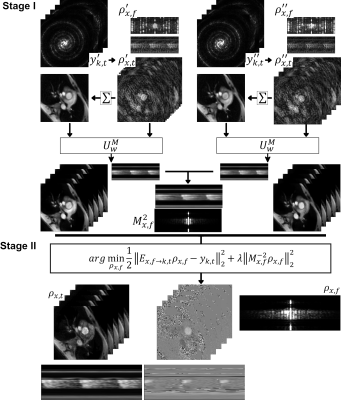 |
Machine Learning aided k-t SENSE for fast reconstruction of highly accelerated PCMR data
Grzegorz Tomasz Kowalik1, Javier Montalt-Tordera1, Jennifer Steeden1, and Vivek Muthurangu1
1Institute of Cardiovascular Science, University College London, London, United Kingdom
The Machine Learning aided k-t SENSE for the reconstruction of highly undersampled GASperturbed PCMR data is validated. We introduce a modified version of the u-net Convolutional Neural Network (u-net M) that utilises the spatial signal distribution information to improve removal of the MR image magnitude aliases. The high resolution magnitude predictions enable creation of regularisation priors used in the k-t SENSE for the final reconstruction of the PCMR data. 20 patients were scanned in the in-vivo validataion. The technique enabled ~3.6x faster processing than the CS reconstruction with no statistical difference in the measured peak mean velocity and stroke volumes.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.