Oral
Artificial Intelligence Applied to MSK MRI
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 4 | 16:00 - 18:00 | Moderators: Fang Liu & Mingrui Yang |
0803. |
Deep Learning-based Semi-supervised Meniscus Segmentation with Uncertainty Estimation
Siyue LI1, Shutian ZHAO1, Yongcheng YAO1, and Weitian CHEN1
1AI in Radiology Laboratory, Department of Imaging and Interventioanl Radiology, The Chinese University of Hong Kong, Hongkong, Hong Kong
Accurate segmentation of the meniscus is valuable for clinical diagnosis and treatment of knee joint diseases. Due to expensive and time-consuming medical image data annotation, it is challenging to obtain sufficient labeled data for deep learning-based segmentation of meniscus. We investigated deep-learning based semi-supervised approaches with uncertainty estimation for meniscus segmentation using MR images.
|
||
0804.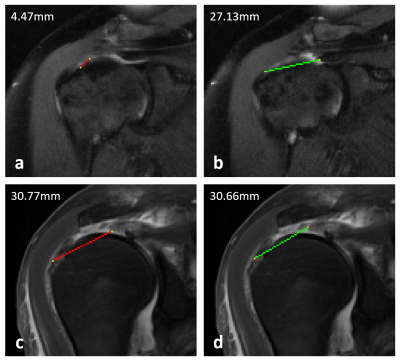 |
Feasibility of deep learning–based automated rotator cuff tear measurements on shoulder MRI
Dana Lin1, Michael Schwier2, Bernhard Geiger2, Esther Raithel3, and Michael Recht1
1NYU Grossman School of Medicine, New York, NY, United States, 2Siemens Medical Solutions USA, Princeton, NJ, United States, 3Siemens Healthcare GmbH, Erlangen, Germany
Rotator cuff tear size is a critical determinant of patient prognosis and surgical outcomes. Radiologists routinely make rotator cuff measurements as part of their MRI interpretation, which can be tedious and subject to variation among readers. This lends itself to a potential application for deep learning to increase efficiency and decrease variability in this task. In this study, we developed a DL model to generate measurements for full-thickness supraspinatus tendon tears.
|
||
0805.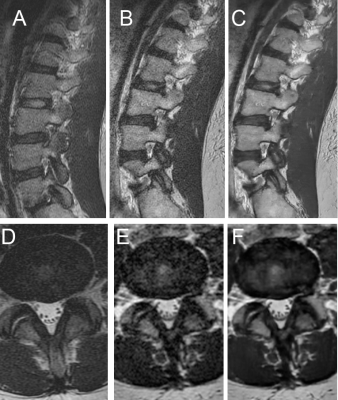 |
Evaluation of Deep-Learning Reconstructed High-Resolution 3D Lumbar Spine MRI to Improve Image Quality
Simon Sun1, Ek Tsoon Tan1, John A Carrino1, Douglas Nelson Mintz1, Meghan Sahr1, Yoshimi Endo1, Edward Yoon1, Bin Lin1, Robert M Lebel2, Suryanarayan Kaushik2, Yan Wen2, Maggie Fung2, and Darryl B Sneag1
1Radiology, Hospital for Special Surgery, New York, NY, United States, 2GE Healthcare, Chicago, IL, United States
Advances in deep-learning algorithms aiming to improve image quality have not yet been well studied for their use in clinical interpretation. In this study, we compared interobserver agreement and image quality for lumbar spine (L-spine) MRI assessment of 3D T2-weighted fast spin echo (T2w-FSE) MRI, with and without deep learning (DLRecon) reconstructions, as well as standard-of-care (SOC) 2D T2w-FSE MRI. This pilot study demonstrated that interobserver agreement for variables of interest was good to very good regardless of reconstruction or sequence type, and overall image quality of DLRecon was not inferior despite significant reduction in scanning time.
|
||
 |
0806.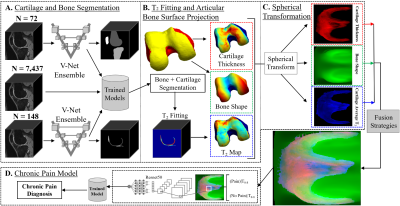 |
DeepPain: Uncovering Associations Between Data-Driven Learned qMRI Biomarkers and Chronic Pain
Alejandro Morales Martinez1, Jinhee Lee1, Francesco Caliva1, Claudia Iriondo1, Sarthak Kamat1, Sharmila Majumdar1, and Valentina Pedoia1
1UCSF, San Francisco, CA, United States
Large-scale analysis of the relationship between learned qMRI biomarkers and chronic knee pain. 7,437 patient timepoints reporting chronic pain were used to train three different deep learning models for bone shape, cartilage thickness, and cartilage T2 biomarkers for the femur, tibia, and patella. The true chronic knee pain predictions for each trained model were further investigated with Grad-CAM and the max activation values for each model were sorted into clinically relevant anatomical compartments for each bone. Bone shape and cartilage T2 seemed to be spatially correlated based on the results of the analysis.
|
|
0807.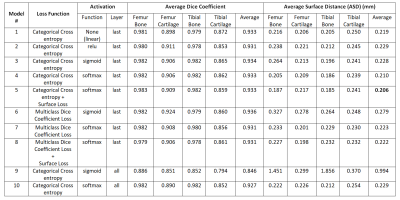 |
The Effect of Activation Functions and Loss Functions on Deep Learning Based Fully Automated Knee Joint Segmentation
Sibaji Gaj1, Dennis Chan1, and Xiaojuan Li1
1Department of Biomedical Engineering, Cleveland Clinic, Cleveland, OH, United States
Studies on systematic evaluations of effects of activation functions and loss functions on deep learning-based automated knee compartments segmentation models are limited. In this work, we present a 2D-UNet model for simultaneous automated bone and cartilage segmentation, and analyze the effect of different activation functions (rectified linear unit[relu], sigmoid and softmax) at all or last layer, and different loss functions (categorical cross-entropy, multiclass dice coefficient loss) with and without surface distance weights, on model performance. The results showed significant performance differences in average surface distance (ASD) between different activation functions. Adding surface distance to loss functions improved segmentation performances.
|
||
0808.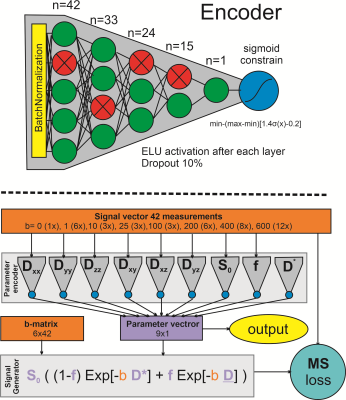 |
Combined IVIM and DTI fitting of muscle DWI data using a self learning physics informed neural network
Martijn Froeling1
1Imaging and oncology, University medical center utrecht, Utrecht, Netherlands
For accurate fitting of muscle diffusion tensor imaging data, many methods have been proposed. In this study, the performance of an unsupervised physics-informed deep learning method for IVIM-DTI fitting of muscle DTI data is investigated. The neural net comprised 9 fully connected networks and was tested on 20 upper leg DWI datasets. It trained in 45s and fitting a full dataset took around 4s. Although the parameter maps of the traditional and NN fitting look similar all parameters were significantly different. The network is capable of fitting the model within seconds but the differences need to be further investigated.
|
||
0809.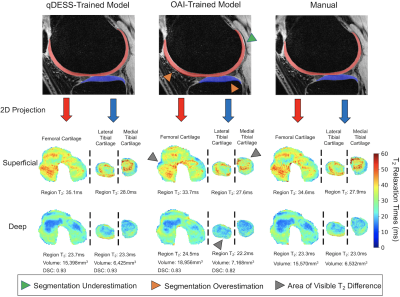 |
Generalizability of Deep-Learning Segmentation Algorithms for Measuring Cartilage and Meniscus Morphology and T2 Relaxation Times
Andrew M Schmidt1, Arjun D Desai1, Lauren E Watkins2, Hollis Crowder3, Elka B Rubin1, Valentina Mazzoli1, Quin Lu4, Marianne Black1,3, Feliks Kogan1, Garry E Gold1,2, Brian A Hargreaves1,5, and Akshay S Chaudhari1,6
1Radiology, Stanford University, Stanford, CA, United States, 2Bioengineering, Stanford University, Stanford, CA, United States, 3Mechanical Engineering, Stanford University, Stanford, CA, United States, 4Philips Healthcare North America, Gainesville, FL, United States, 5Electrical Engineering, Stanford University, Stanford, CA, United States, 6Biomedical Data Science, Stanford University, Stanford, CA, United States
Automated segmentation using deep-learning can expedite segmentation tasks, but algorithm generalizability to unseen datasets is unknown. Here, we used two knee segmentation algorithms, each trained separately on Osteoarthritis Initiative double-echo steady-state (DESS) scans and quantitative DESS (qDESS) scans, to segment cartilage and meniscus from qDESS datasets from four independent studies. We compared manual-vs-automatic segmentation accuracy for morphology and T2 map variations. We show that OAI-DESS-trained models may be suitable for quantifying relaxometry parameters in qDESS datasets but likely require fine-tuning to accurately quantify cartilage morphology. In contrast, qDESS-trained models generalize well to additional qDESS datasets for both morphology and T2.
|
||
 |
0810.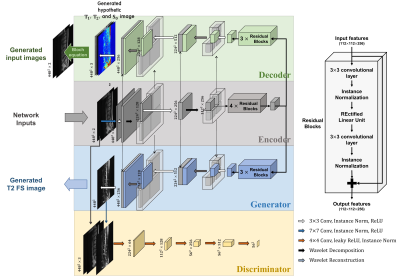 |
Generative Adversarial Network for T2-Weighted Fat Saturation MR Image Synthesis Using Bloch Equation-based Autoencoder Regularization
Sewon Kim1, Hanbyol Jang2, Seokjun Hong2, Yeong Sang Hong3, Won C. Bae4,5, Sungjun Kim*3,6, and Dosik Hwang*2
1Electrical and electronic engineering, Yonsei University, Seoul, Korea, Republic of, 2Yonsei University, Seoul, Korea, Republic of, 3Gangnam Severance Hospital, Seoul, Korea, Republic of, 4Department of Radiology, University of California-San Diego, San Diego, CA, United States, 5Department of Radiology, VA San Diego Healthcare System, San Diego, CA, United States, 6Yonsei University College of Medicine, Seoul, Korea, Republic of
We proposed a Bloch equation-based autoencoder regularization Generative Adversarial Network (BlochGAN) to generate T2-weighted fat saturation (T2 FS) images from T1-weighted (T1-w) and T2-weighted (T2-w) images for spine diagnosis. Our method can reduce the cost for acquiring multi-contrast images by reducing the number of contrasts to be scanned. BlochGAN properly generates the target contrast images by using GAN trained with the autoencoder regularization based on bloch equation, which is the basic principal of MR physics for identifying the physical basis of the contrasts. Our results demonstrate that BlochGAN achieved quantitatively and qualitatively superior performance compared to conventional methods.
|
|
0811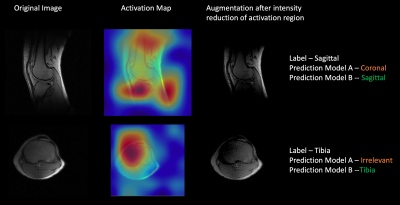 |
Data augmentation using features from activation maps improved performance for deep learning based automated knee prescription Video Permission Withheld
Deepa Anand1, Dattesh Shanbhag1, Preetham Shankpal1, Chitresh Bhushan2, Desmond Teck Beng Yeo2, Thomas K Foo2, and Radhika Madhavan2
1GE Healthcare, Bangalore, India, 2GE Global Research, Niskayuna, NY, United States
Data augmentation techniques have been routinely used in computer vision for simulating variations in input data and avoid overfitting. Here we propose a novel method to generate simulated images using features derived from activation maps of a deep neural network, which could mimic image variations due to MRI acquisition and hardware. Gradient-weighted Class Activation Mappings were used to identify regions important to classification output, and generate images with these regions obfuscated to mimic adversarial scenarios relevant for imaging variations. Training with images using the proposed data augmentation framework resulted in improved accuracy and enhanced robustness of knee MRI image classification.
|
||
0812.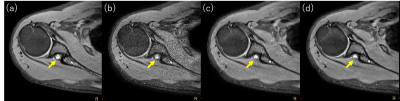 |
Ultrafast motion-minimized shoulder MRI with a deep learning constrained Compressed SENSE reconstruction
Jihun Kwon1, Masami Yoneyama1, Takashige Yoshida2, Kohei Yuda2, Yuki Furukawa2, Johannes M. Peeters3, and Marc Van Cauteren3
1Philips Japan, Tokyo, Japan, 2Tokyo Metropolitan Police Hospital, Nakano, Japan, 3Philips Healthcare, Best, Netherlands Shoulder MRI is typically acquired with multiple number of signals averaged (NSA) in order to average out breathing motion artifacts. However, higher NSA leads to a longer scan time and patient discomfort. In this study, we investigated the use of a deep learning-based reconstruction algorithm to highly accelerate shoulder MRI. Adaptive-CS-Net, a deep neural network previously introduced at the 2019 fastMRI challenge, was expanded and presented here as a Compressed-SENSE Artificial Intelligence (CS-AI) reconstruction. The purpose of this study was to compare the image quality of shoulder MRI between reference and accelerated methods; SENSE, Compressed-SENSE, and CS-AI. |
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.