Oral
Data Processing & Software Tools
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 1 | 14:00 - 16:00 | Moderators: Mathieu Boudreau & Eve LoCastro |
 |
0725.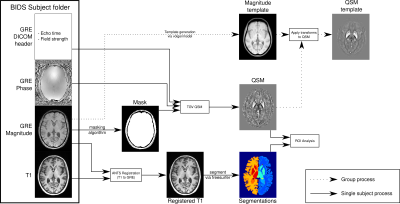 |
QSMxT - A cross-platform, flexible, lightweight, and scalable processing pipeline for quantitative susceptibility mapping
Ashley Stewart1,2, Simon Daniel Robinson2,3,4, Kieran O'Brien1,2,5, Jin Jin1,2,5, Angela Walls6, Aswin Narayanan2, Markus Barth1,2,7, and Steffen Bollmann1,2,7
1Centre for Innovation in Biomedical Imaging Technology, University of Queensland, Brisbane, Australia, 2Centre for Advanced Imaging, University of Queensland, Brisbane, Australia, 3High Field MR Center, Department of Biomedical Imaging and Image-Guided Therapy, Medical University of Vienna, Vienna, Austria, 4Department of Neurology, Medical University of Graz, Graz, Austria, 5Siemens Healthcare Pty Ltd, Brisbane, Australia, 6Clinical & Research Imaging Centre, South Australian Health and Medical Research Institute, Adelaide, Australia, 7School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia
We developed an open-source QSM processing framework, QSMxT, that provides a full QSM workflow including converting DICOM data to BIDS, a variety of robust masking strategies, phase unwrapping, background field correction, dipole inversion and region-of-interest analyses based on automated anatomical segmentations. We make all required external dependencies available in a reproducible and scalable analysis environment enabling users to process QSM data for large groups of participants on any operating system in a robust way.
|
|
 |
0726. |
Physiopy: A community-driven suite of tools for physiological recordings in neuroimaging
Katherine Louise Bottenhorn1, Daniel Alcalà-Lopez2, Apoorva Ayyagari3, Molly G Bright4, César Caballero-Gaudes2, Inés Chavarria2, Vicente Ferrer2, Soichi Hayashi5, Vittorio Iacovella6, François Lespinasse7, Ross Davis Markello8, Stefano Moia2, Robert Oostenveld9,10,
Taylor Salo1, Rachael Stickland4, Eneko Uruñuela2, Merel Margaretha van der Thiel11, and Kristina M Zvolanek12
1Department of Psychology, Florida International University, Miami, FL, United States, 2Basque Center on Cognition, Brain and Language, Donostia, Spain, 3Northwestern University, Chicago, IL, United States, 4Physical Therapy and Human Movement Sciences, Northwestern University, Chicago, IL, United States, 5Indiana University, Bloomington, IN, United States, 6CIMeC - Center for Mind / Brain Sciences, The University of Trento, Trento, Italy, 7Psychology, Université de Montréal, Montréal, QC, Canada, 8McGill University, Montréal, QC, Canada, 9Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands, 10NatMEG, Karolinska Institutet, Stockholm, Sweden, 11Department of Radiology & Nuclear Medicine, Maastricht University Medical Center, Maastricht, Netherlands, 12Biomedical Engineering, Northwestern University, Chicago, IL, United States
Physiopy is a community-driven effort that aims to offer a full data preparation pipeline for non-neural physiological recordings in neuroimaging research and to build consensus on “best practices” among researchers of this specific domain. Its primary goal is to facilitate physiological data collection and sharing, according to an existing data standard and ontology. Corollary goals of the community are to provide recommendations and tools for (1) physiological data acquisition in the MR environment, (2) appropriate processing of these data, (3) organization of resulting datasets and meta-data, and (4) computing metrics for the removal of confounding physiological signals from fMRI data.
|
|
0727.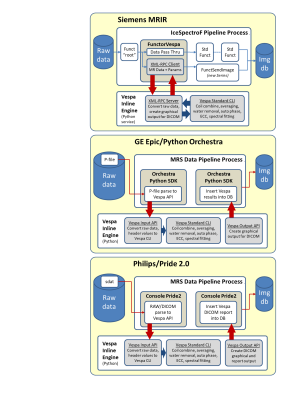 |
Across-vendor, inline standardized spectral analysis for single voxel MRS data acquisition at 3T
Brian J Soher1, Dinesh K Deelchand2, Sandeep Ganji3, Ralph Noeske4, Adam Berrington5, James Joers2, and Gulin Oz2
1Radiology, Duke University Medical Center, Durham, NC, United States, 2University of Minnesota, Minneapolis, MN, United States, 3Philips Healthcare, Rochester, MN, United States, 4GE Healthcare, Berlin, Germany, 5University of Nottingham, Nottingham, United Kingdom
The minimal spectral analysis methods described by MRS community consensus are not available in manufacturers’ MRS software. MRS data is still transferred off the scanner for advanced analysis leading to more variability in MRS results and impediments for adoption within a clinical DICOM workflow. We describe the Vespa Inline Engine (VIE), a flexible add-in module for the open-source Vespa spectral analysis package. VIE provides equivalent, inline MRS processing on GE, Siemens and Philips platforms. Raw MRS data sent to the VIE analysis returns results to the DICOM workflow as graphical and tabular DICOM images.
|
||
 |
0728.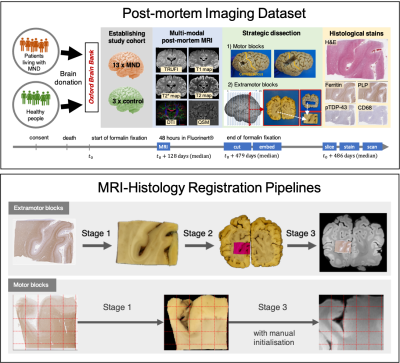 |
TIRL: Automating Deformable Slice-to-Volume Registration Between Stand-Alone Histology Sections and Post-Mortem MRI
Istvan N Huszar1, Karla L Miller1, and Mark Jenkinson1
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
An open-source framework (TIRL) and a novel 3-stage pipeline are presented to automate the registration between sparsely sampled small histology sections and 3-D MRI data for validating imaging biomarkers. In addition to affine registration, deformable slice-to-volume registration is employed to compensate for both in-plane and through-plane distortions of the histology sections. Each stage of the pipeline is shown to achieve submillimetre precise alignments, surpassing the accuracy of previous methods. With photographic intermediaries the pipeline is fully automatic and does not depend on serial histological sectioning or specialist cutting and stain automation hardware. The tools are provided as part of FSL.
|
|
 |
0729.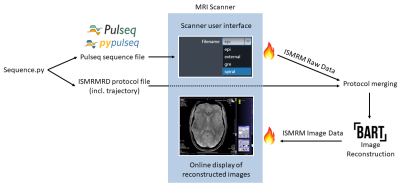 |
Open-Source MR Imaging Workflow
Marten Veldmann1, Philipp Ehses1, Kelvin Chow2, Maxim Zaitsev3, and Tony Stöcker1,4
1MR Physics, German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany, 2MR R&D Collaborations, Siemens Medical Solutions USA Inc., Chicago, IL, United States, 3Department of Radiology - Medical Physics, University Medical Center Freiburg, Freiburg, Germany, 4Department of Physics & Astronomy, University of Bonn, Bonn, Germany
We demonstrate an open-source MR imaging workflow for MR sequence development and online image reconstruction. The open-source approach improves vendor-independent reproducibility of new or modified MR sequences and reconstruction algorithms. An extensible online reconstruction pipeline allows for fast evaluation of the acquired data on the scanner directly. The pipeline supports implementation of advanced open-source image reconstruction techniques and is not restricted to a specific sequence. We successfully implemented an open-source spiral sequence with Pulseq for execution at a 7T MR scanner. A non-Cartesian image reconstruction was done with the BART toolbox using GIRF trajectory prediction.
|
|
0730.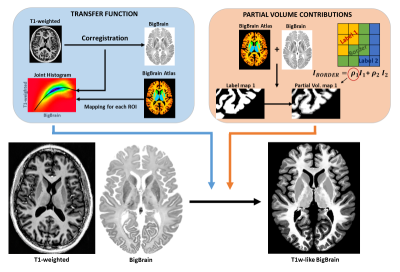 |
BigBrain-MR: a computational phantom for ultra-high-resolution MR methods development
Cristina Sainz Martinez1,2, Mathieu Lemay1, Meritxell Bach Cuadra2,3,4, and João Jorge1,2
1Systems Division, Swiss Center for Electronics and Microtechnology (CSEM), Nêuchatel, Switzerland, 2Medical Image Analysis Laboratory (MIAL), Center for Biomedical Imaging (CIBM), Lausanne, Switzerland, 3Signal Processing Laboratory (LTS5), École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 4Department of Radiology, Centre Hospitalier Universitaire Vaudois (CHUV) and University of Lausanne (UNIL), Lausanne, Switzerland
With the increasing importance of ultra-high field systems, suitable simulation platforms are needed for the development of high-resolution imaging methods. Here, we propose a realistic computational brain phantom at 100μm resolution, by mapping fundamental MR properties (e.g., T1, T2, coil sensitivities) from existing brain MRI data to the fine-scale anatomical space of BigBrain, a publicly-available 100μm-resolution ex-vivo image obtained with optical methods. We propose an approach to map image contrast from lower-resolution MRI data to BigBrain, retaining the latter’s fine structural detail. We then show its value for methodological development in two applications: super-resolution, and reconstruction of highly-undersampled k-space acquisitions.
|
||
 |
0731.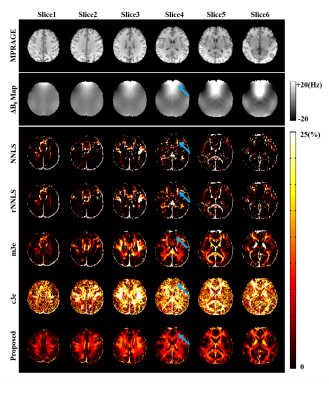 |
Improved Estimation of Myelin Water Fractions with Learned Parameter Distributions
Yudu Li1,2, Jiahui Xiong1,2, Rong Guo1,2, Yibo Zhao1,2, Yao Li3,4, and Zhi-Pei Liang1,2
1Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL, United States, 3School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 4Med-X Research Institute, Shanghai, China
Myelin water fraction (MWF) mapping can substantially improve our understanding of several demyelinating diseases. While MWF maps can be obtained from multi-exponential fitting of multi-echo imaging data, current solutions are often very sensitive to noise and modeling errors. This work addresses this problem using a new model-based method. This method has two key novel features: a) an improved signal model capable of compensating practical signal errors, and b) incorporation of parameter distributions and low-rank signal structures. Both simulation and experimental results show that the proposed method significantly outperforms the conventional methods currently used for MWF estimation.
|
|
 |
0732.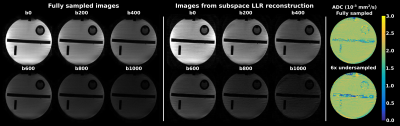 |
Spatiotemporal encoding for DWI of brain and prostate using subspace-constrained sampling and locally-low-rank regularized reconstruction
Martins Otikovs1, Ankit Basak1, and Lucio Frydman1
1Weizmann Institute of Science, Rehovot, Israel
The benefits of performing locally low-rank (LLR) reconstruction on subsampled diffusion weighted (DW) data employing spatiotemporal encoding (SPEN) methods, is investigated. SPEN allows for self-referenced correction of motion-induced phase errors in case of interleaved DW acquisitions, and allows to overcome distortions otherwise observed along EPI’s phase-encoded dimension. In combination with LLR regularization and with joint subsampling of b-weighted and interleaved images, additional improvements are demonstrated. The method is tested on phantoms, and improvements in in-vivo brain and prostate scans on volunteers are demonstrated.
|
|
0733.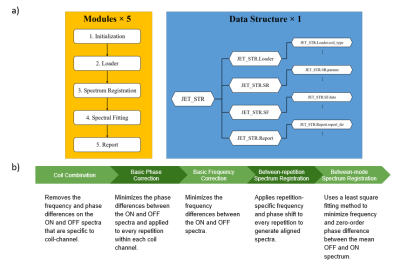 |
JET - A Matlab toolkit for automated J-difference-edited MR spectra processing of in vivo mouse MEGA-PRESS study at 9.4T
Chen Liu1, David Jing Ma2, Nanyan Zhu3, Kay Igwe2, Jochen Weber2, Roshell Li4, Emily Turner Wood5, Wafae Labriji6, Vasile Stupar6, Yanping Sun7, Neil Harris8, Antoine Depaulis6, Florence Fauvelle 9, Scott A. Small10,11,12,
Douglas L. Rothman13, and Jia Guo10,14
1Department of Electrical Engineering and the Taub Institute, Columbia University, New York, NY, United States, 2Columbia University, New York, NY, United States, 3Department of Biological Sciences and the Taub Institute, Columbia University, New York, NY, United States, 4Department of Biomedical Engineering, Columbia University, New York, NY, United States, 5University of California, Los Angeles, Los Angeles, CA, United States, 6Grenoble Institut Neurosciences (GIN), Grenoble, France, 7Herbert Irving Comprehensive Cancer Centre, Columbia University, New York, NY, United States, 8Department of Neurosurgery, University of California, Los Angeles, Los Angeles, CA, United States, 9Grenoble MRI Facility IRMaGe, France, France, 10Department of Psychiatry, Columbia University, New York, NY, United States, 11Department of Neurology, Columbia University, New York, NY, United States, 12Taub Institute for Research on Alzheimer's Disease and the Aging Brain, Columbia University, New York, NY, United States, 13Radiology and Biomedical Imaging and of Biomedical Engineering, Yale University, New Haven, CT, United States, 14Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States
Spectral editing studies in mice brains have been limited due to difficulty in spectrum processing and lack of software package analysis. However, in preclinical studies, mouse models play an important role in understanding effects of drugs and its impact on the nervous system. JET is a fully automated software that performs raw data conversion, spectrum registration, spectral quality assessment and metabolite quantification of MEGA-PRESS mouse data at 9.4T. In this work, we first introduce the automated spectra processing pipeline of JET and further demonstrate its utilities in mouse studies.
|
||
0734.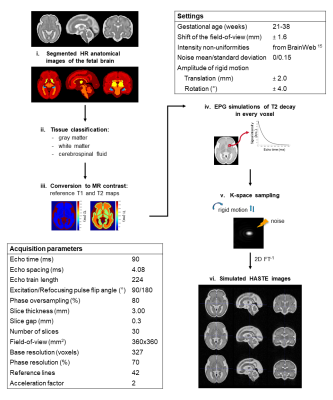 |
A Magnetic Resonance Imaging Simulation Framework of the Developing Fetal Brain
Hélène Lajous1,2, Tom Hilbert1,3,4, Christopher W. Roy1, Sébastien Tourbier1, Priscille de Dumast1,2, Yasser Alemán-Gómez1, Thomas Yu4, Patric Hagmann1, Mériam Koob1, Vincent Dunet1, Tobias Kober1,3,4, Matthias Stuber1,2, and Meritxell Bach Cuadra1,2,4
1Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 2CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 3Advanced Clinical Imaging Technology (ACIT), Siemens Healthcare, Lausanne, Switzerland, 4Signal Processing Laboratory 5 (LTS5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
Accurate characterization of in utero human brain maturation is critical. However, the limited number of exploitable magnetic resonance acquisitions not corrupted by motion in this cohort of sensitive subjects hinders the validation of advanced image processing techniques. Numerical simulations can mitigate these limitations by providing a controlled environment with a known ground truth. We present a flexible framework that simulates magnetic resonance acquisitions of the fetal brain in a realistic setup including stochastic motion. From simulated images comparable to clinical acquisitions, we assess the accuracy and robustness of super-resolution fetal brain magnetic resonance imaging with respect to noise and motion.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.