Oral
Diffusion: Encoding & Estimation
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 3 | 18:00 - 20:00 | Moderators: Guillaume Gilbert & Sjoerd Vos |
 |
0397.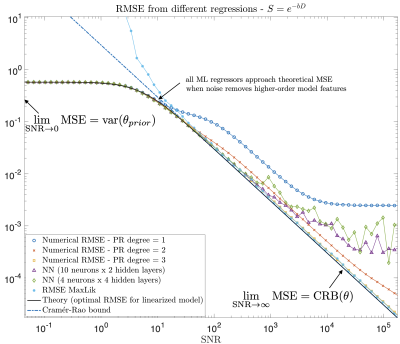 |
How do we know we measure tissue parameters, not the prior?
Santiago Coelho1, Els Fieremans1, and Dmitry S. Novikov1
1Center for Biomedical Imaging and Center for Advanced Imaging Innovation and Research (CAI$^2$R), Department of Radiology, New York University School of Medicine, New York, NY, United States
In the machine-learning (ML) era, we are transitioning from max-likelihood parameter estimation to learning the mapping from measurements to model parameters. While such maps look smooth, there is danger of them becoming too smooth: At low SNR, ML estimates become the mean of the training set. Here we derive fit quality (MSE) as function of SNR, and show that MSE for various ML methods (regression, neural-nets, random forest) approaches a universal curve interpolating between Cramér-Rao bound at high SNR, and variance of the prior at low SNR. Theory is validated numerically and on white matter Standard Model in vivo.
|
|
 |
0398.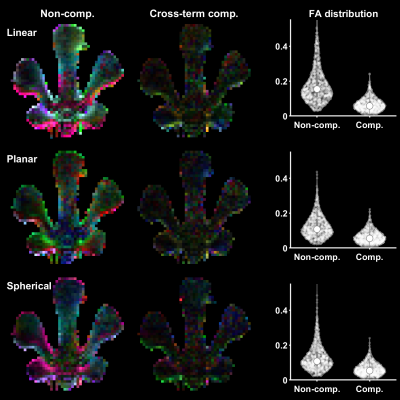 |
Gradient waveform design for cross-term-compensated diffusion MRI: Demonstration of tensor-valued encoding in phantom and simulations
Filip Szczepankiewicz1 and Jens Sjölund2
1Clinical Sciences Lund, Lund University, Lund, Sweden, 2Department of Information Technology, Uppsala University, Uppsala, Sweden
Diffusion weighted imaging is perturbed by the presence of background gradients, or so-called 'cross-terms,' causing bias in estimated parameters and fiber orientations. In this work, we present a novel gradient waveform design that removes the cross-term sensitivity entirely. This design is valuable for diffusion MRI methods that are otherwise corrupted by background gradients, and it also facilitates arbitrary sequence timing, b-tensor shapes and suppression of concomitant gradient effects.
|
|
 |
0399.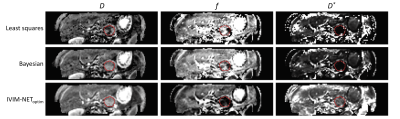 |
Improved unsupervised physics-informed deep learning for intravoxel-incoherent motion modeling
Misha P. T. Kaandorp1,2,3, Sebastiano Barbieri4, Remy Klaassen5, Hanneke W.M. van Laarhoven5, Hans Crezee6, Peter T. While2,3, Aart J. Nederveen1, and Oliver J. Gurney-Champion1
1Department of Radiology and Nuclear Medicine, Amsterdam UMC, Amsterdam, Netherlands, 2Department of Radiology and Nuclear Medicine, St. Olav's University Hospital, Trondheim, Norway, 3Department of Circulation and Medical Imaging, NTNU: Norwegian University of Science and Technology, Trondheim, Norway, 4Centre for Big Data Research in Health, UNSW, Sydney, Australia, 5Department of Medical Oncology, Amsterdam UMC, Amsterdam, Netherlands, 6Department of Radiation Oncology, Amsterdam UMC, Amsterdam, Netherlands
We implemented an improved unsupervised physics-informed deep neural network approach for intravoxel-incoherent motion modeling to DWI data by exploring several hyperparameters. Whereas the original IVIM-NETorig showed high dependency between the predicted IVIM parameters, our optimized approach resolved this high dependency, produced better accuracy and was more consistent. In simulations, IVIM-NEToptim outperformed least-squares and Bayesian fitting approaches. In patients with pancreatic ductal adenocarcinoma, IVIM-NEToptim produced substantially less noisy parameter maps and lower intersession within-subject standard deviations than the alternatives. IVIM-NEToptim also detected the most individual patients with significant parameter changes in the group of patients who received chemoradiotherapy.
|
|
0400.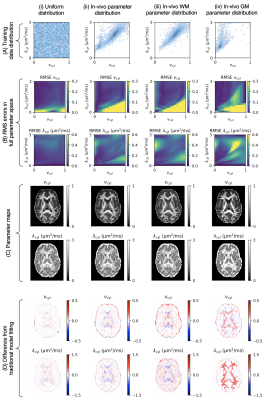 |
Training Data Distribution Significantly Impacts the Estimation of Tissue Microstructure with Machine Learning
Noemi G. Gyori1,2, Marco Palombo1, Christopher A. Clark2, Hui Zhang1, and Daniel C. Alexander1
1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Great Ormond Street Institute of Child Health, University College London, London, United Kingdom
The performance of supervised machine learning tools is only as good as the data used to train them. In this work, we investigate the impact of training data distribution on tissue microstructure estimates in the human brain. We focus on two strategies: uniform sampling from the entire parameter space and sampling from parameter combinations observed using traditional model fitting. We demonstrate that training on previously observed combinations may be advantageous for detecting small variations in healthy tissue. However, for detecting atypical tissue abnormalities, our results favour uniform training data sampling in which all plausible parameter combinations are represented.
|
||
0401.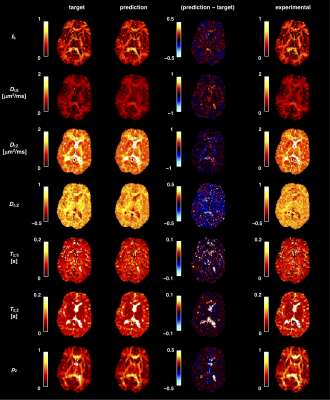 |
On the use of neural networks to fit high-dimensional microstructure models
João Pedro de Almeida Martins1,2, Markus Nilsson1, Björn Lampinen3, Marco Palombo4, Carl-Fredrik Westin5,6, and Filip Szczepankiewicz1,5,6
1Department of Clinical Sciences, Radiology, Lund University, Lund, Sweden, 2Department of Radiology and Nuclear Medicine, St. Olav's University Hospital, Trondheim, Norway, 3Department of Clinical Sciences, Medical Radiation Physics, Lund University, Lund, Sweden, 4Centre for Medical Image Computing and Dept of Computer Science, University College London, London, United Kingdom, 5Radiology, Brigham and Women’s Hospital, Boston, MA, United States, 6Harvard Medical School, Boston, MA, United States
The application of function fitting neural networks in microstructural MRI has so far been restricted to lower-dimensional biophysical models. Moreover, the data sufficiency requirements of learning-based approaches remain unclear. Here, we use supervised learning to vastly accelerate the fitting of a high-dimensional relaxation-diffusion model of tissue microstructure and develop analysis tools for assessing the accuracy and sensitivity of model fitting networks. The developed learning-based fitting pipelines were tested on relaxation-diffusion data acquired with optimal and sub-optimal protocols. We found no evidence that machine-learning algorithms can correct for a degenerate fitting landscape or replace a careful design of the acquisition protocol.
|
||
 |
0402.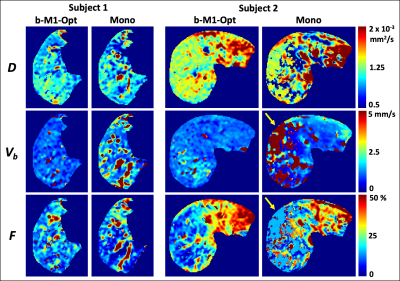 |
b-M1-Optimized Waveforms for Improved Stability of Quantitative Intravoxel Incoherent Motion DWI
Gregory Simchick1,2, Ruiqi Geng1,2, Yuxin Zhang1,2, and Diego Hernando1,2,3
1Radiology, University of Wisconsin-Madison, Madison, WI, United States, 2Medical Physics, University of Wisconsin-Madison, Madison, WI, United States, 3Biomedical Engineering, University of Wisconsin-Madison, Madison, WI, United States
Multiple studies have demonstrated the potential utility of intravoxel incoherent motion (IVIM) DWI in the evaluation of liver disease. However, IVIM estimates obtained using conventional monopolar diffusion gradient waveforms often suffer from high variability and instability. In this work, Cramer-Rao lower bound (CRLB) optimization was performed to determine the optimal sampling of b-M1-space based on noise performance. Then, based on this optimized sampling, b-M1-optimized waveforms were designed and employed in order to obtain IVIM estimates with improved stability in the right lobe of the liver in volunteers in comparison to conventional monopolar waveforms.
|
|
 |
0403.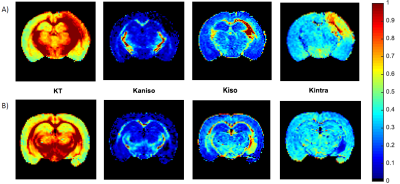 |
Resolving the underlying sources of diffusion kurtosis in focal ischemia by Correlation Tensor MRI
Rita Alves1, Rafael Neto Henriques1, Sune Nørhøj Jespersen2,3, and Noam Shemesh1
1Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 2Center of Functionally Integrative Neuroscience (CFIN) and MINDLab, Department of Clinical Medicine, Aarhus University, Aarhus, Denmark, 3Department of Physics and Astronomy, Aarhus University, Aarhus, Denmark
Correlation Tensor MRI (CTI) has been recently introduced for resolving the underlying sources of diffusional kurtosis. Here, we aimed to resolve the underlying kurtosis sources in ischemic tissue. Ex and in vivo CTI experiments in a mouse model of ischemia revealed enhanced sensitivity and specificity compared to their conventional counterparts. Our results suggest that microscopic kurtosis – associated with restricted diffusion and structural disorder – substantially contributes to the total kurtosis excess likely reflecting excitotoxic properties. Kurtosis associated with diffusion magnitude variance better reflected edema and free water. These first results are promising for elucidating biological factors in ischemia.
|
|
 |
0404.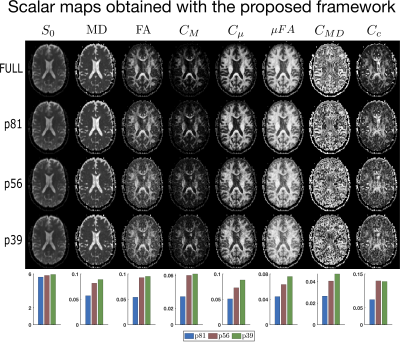 |
Enforcing positivity constraints in Q-space Trajectory Imaging (QTI) allows for reduced scan time
Deneb Boito1,2, Magnus Herberthson3, Tom Dela Haije4, and Evren Özarslan1,2
1Department of Biomedical Engineering, Linköping University, Linköping, Sweden, 2Center for Medical Image Science and Visualization, Linköping University, Linköping, Sweden, 3Department of Mathematics, Linköping University, Linköping, Sweden, 4Department of Computer Science, University of Copenhagen, Copenhagen, Denmark
We explore the feasibility of reducing scan time while retaining interpretable results when analysing multidimensional diffusion MRI data. The methods commonly employed for the estimation of the QTI model parameters might suffer severely from data under-sampling, making their use limited in clinical practice where acquisition time is limited. We show that by imposing relevant positivity constraints in the estimation of the mean, covariance, and moment tensors, the QTI+ framework strongly improves the quality of the derived maps, particularly so for data acquisition protocols featuring few number of volumes.
|
|
 |
0405.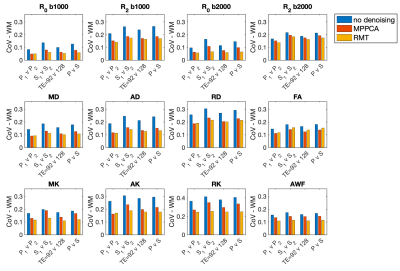 |
Random matrix theory denoising minimizes cross-scanner,-protocol variability and maximizes repeatability of higher-order diffusion metrics
Benjamin Ades-Aron1, Santiago Coelho1, Jelle Veraart1, Gregory Lemberskiy1, Genevieve Barroll1, Steven Baete1, Timothy Shepherd1, Dmitry S. Novikov1, and Els Fieremans1
1Radiology, NYU School of Medicine, New York, NY, United States
Translation of diffusion MRI-derived quantitative biomarkers into clinical decision making has been hampered by within-scanner and cross-scanner variability. We compare intra-scan, cross-scan, and cross-protocol variability of multi-shell diffusion MRI for three subjects and evaluated two Random Matrix Theory (RMT)-based denoising techniques to enhance repeatability. Without denoising, best scan-rescan repeatability was found for intra-scanner measurements with the highest SNR (shortest TE), RMT-based denoising greatly reduced variability across scanners and TE, resulting in coefficients of variation about 5% for all comparisons. Increased precision across scanners and protocols should increase statistical power and further enable clinical trials of quantitative higher-order diffusion MRI.
|
|
0406.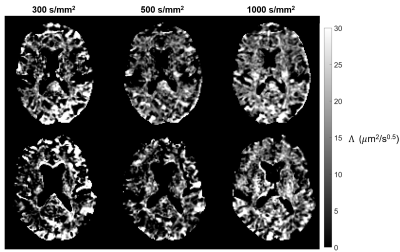 |
A time-efficient OGSE sequence with spiral readout for an improved depiction of diffusion dispersion
Eric Seth Michael1, Franciszek Hennel1, and Klaas Paul Pruessmann1
1Institute for Biomedical Engineering, ETH Zurich and University of Zurich, Zurich, Switzerland
Oscillating gradient spin-echo (OGSE) sequences typically suffer low diffusion sensitization and long echo times, thereby compromising the diffusion-to-noise ratio (DNR). To address these issues, recently proposed OGSE shapes providing increased diffusion sensitization were combined with single-shot spiral readout trajectories using a high-performance gradient system. This implementation was used to study the frequency dependence of diffusivity in the in-vivo human brain for OGSE frequencies up to 125 Hz and b-values up to 1000 s/mm2, yielding an improved depiction of diffusion dispersion. The results indicate the significance of higher b-values in characterizing this relationship.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.