Oral
Functional Connectivity Across Species
ISMRM & SMRT Annual Meeting • 15-20 May 2021

| Concurrent 3 | 16:00 - 18:00 | Moderators: Pinar Ozbay & Suzanne T Witt |
 |
0569.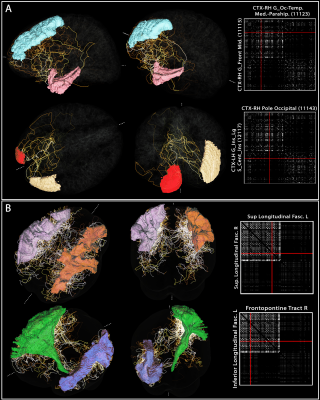 |
Whole-brain vascular connectome: a new approach to investigate the functional brain networks using large-scale angioarchitecture
Michael Bernier1,2, Jingyuan E Chen1,2, Olivia Viessmann1,2, Nina E Fultz1,3, Maxime Chamberland4, Rebecca K Leaf5, Lawrence L Wald1,2,6, and Jonathan R Polimeni1,2,6
1Department of Radiology, A. A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, United States, 2Radiology, Harvard Medical School, Boston, MA, United States, 3Department of Engineering, University of Boston, Boston, MA, United States, 4Cardiff University Brain Research Imaging Centre (CUBRIC), Cardiff, United Kingdom, 5Division of Hematology, Massachusetts General Hospital, Boston, MA, United States, 6Division of Health Sciences and Technology, Massachusetts Institute of Technology, Boston, MA, United States
Blood vessels influence nearby fMRI signals, and patterns of vascular anatomy partly shape the patterns of the BOLD response. To better understand the relationship between large-scale brain networks and vascular anatomy, here we developed an approach for estimating the topology of the vascular network and quantifying how vessel pathways connect between brain regions. We used a blood-pool contrast agent to enhance the vessels, and developed a new method for vessel tracking similar to what is conventionally used to estimate structural connectivity in diffusion MRI. We demonstrate an application this framework by estimating vascular connectivity matrices for the human brain.
|
|
 |
0570.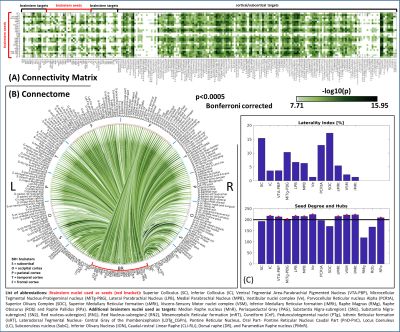 |
Functional connectome of autonomic, limbic, nociceptive, and sensory brainstem nuclei using 7 Tesla resting-state fMRI in living humans
Simone Cauzzo1,2, Kavita Singh2, Matthew Stauder2, Maria Guadalupe Garcia Gomar2, Nicola Vanello3, Claudio Passino1,4, Jeffrey Staab5,6, Iole Indovina7,8, and Marta Bianciardi2
1Life Sciences Institute, Sant'Anna School of Advanced Studies, Pisa, Italy, 2Brainstem Imaging Laboratory, Department of Radiology, Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States, 3Dipartimento di Ingegneria dell’Informazione, University of Pisa, Pisa, Italy, 4Fondazione Toscana Gabriele Monasterio, Pisa, Italy, 5Department of Psychiatry and Psychology, Mayo Clinic, Rochester, MN, United States, 6Department of Otorhinolaryngology – Head and Neck Surgery, Mayo Clinic, Rochester, MN, United States, 7Department of Biomedical and Dental Sciences and Morphofunctional Imaging, University of Messina, Messina, Italy, 8Laboratory of Neuromotor Physiology, IRCCS Santa Lucia Foundation, Roma, Italy
With the goal of bridging the gap between cortical and subcortical (e.g. brainstem) functional connectomes and of defining a baseline for clinical studies, we computed the resting-state functional connectivity maps of 15 brainstem nuclei involved in central autonomic, limbic, nociceptive and sensory functions. We used images acquired with 7 Tesla MRI on a group of 20 healthy subjects and a recently generated probabilistic atlas of brainstem nuclei. We obtained highly significant and stable links with a good overlap with the literature. Our results also provide favorable results on the translatability of our brainstem connectome approach to conventional 3 Tesla data-sets.
|
|
 |
0571.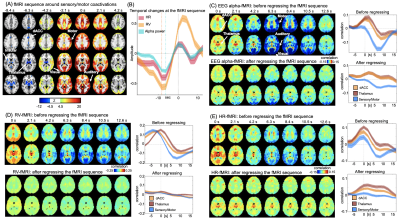 |
Salience network modulation leads a sequence of brain activity that causes resting-state fMRI correlations with EEG and physiological signals
Yameng Gu1, Feng Han1, Lucas Eugene Sainburg1, and Xiao Liu1
1Pennsylvania State Universitya, University Park, PA, United States
Correlations of resting-state fMRI (rsfMRI) with various neural and physiological signals have been observed and interpreted as distinct sources of contributions. Here, we found these rsfMRI-EEG and rsfMRI-physio correlations are similarly characterized by distinct network patterns at different time lags. A similar sequence of fMRI changes led by the salience network modulation is found in resting-state fMRI signals, and removing this sequence significantly diminished the rsfMRI-EEG and rsfMRI-physiology correlations. The results suggest that the rsfMRI correlations with the EEG alpha power and physiological signals originate from a sequence of brain dynamics led by salience network changes.
|
|
 |
0572.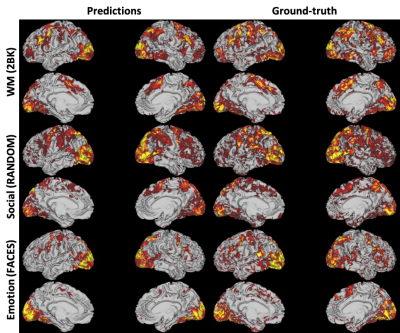 |
Resting-state fMRI Predicts Task Activation Patterns Using a Graph Convolutional Network
Zhangxuan Hu1,2, Hua Guo2, Lihong Wang3, Bing Wu1, and Xue Zhang4
1GE Healthcare, MR Research China, Beijing, China, 2Center for Biomedical Imaging Research, Department of Biomedical Engineering, School of Medicine, Tsinghua University, Beijing, China, 3Department of Psychiatry, University of Connecticut School of Medicine, Farmington, MI, United States, 4Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, United States
Resting-state fMRI has the clinical potential as diagnostic and prognostic markers because of its easy implementation/standardization in data acquisitions, and its ability to parcellate functionally connected neural networks. It is of importance to examine whether the task-free spontaneous activity could be used to predict individuals’ task-induced activation. Here we proposed a graph convolutional network-based framework which utilized the information of the brain connections for the convolution step, and showed the ability of using resting-state fMRI to predict individual differences in activations of tasks from human connectome project. This framework could be extended to other resting-state fMRI researches.
|
|
 |
0573.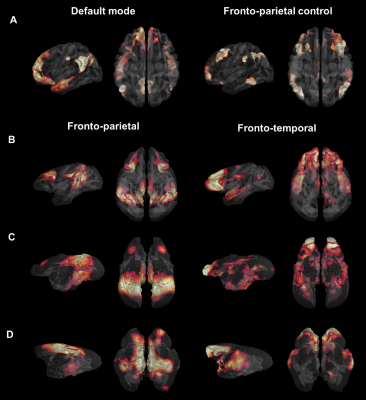 |
Evolutionary gap of the default mode network organization between non-hominid primates and humans
Clément M. Garin1, Yuki Hori2, Stefan Everling 2,3, Christopher T. Whitlow 4, Finnegan J. Calabro 5, Beatriz Luna5, Marc Dhenain 6,7, and Christos Constantinidis 1,8
1Neurobiology and Anatomy, Wake Forest University, Winston Salem, NC, United States, 2Centre for Functional and Metabolic Mapping, Robarts Research Institute, University of Western Ontario, London, ON, Canada, 3Department of Physiology and Pharmacology, The University of Western Ontario, London, ON, Canada, 4Department of Radiology, Section of Neuroradiology, Wake Forest University, Winston Salem, NC, United States, 5Department of Psychiatry, University of Pittsburgh, Pittsburgh, PA, United States, 6Neurodegenerative Diseases Laboratory, Centre National de la Recherche Scientifique (CNRS), Université Paris-Sud, Université Paris-Saclay UMR 9199, Fontenay-aux-Roses, France, 7Commissariat à l’Energie Atomique et aux Energies Alternatives (CEA), Direction de la Recherche Fondamentale (DRF), Institut François Jacob, MIRCen, Fontenay-aux-Roses, France, 8Department of Biomedical Engineering, Vanderbilt University, Nashville, TN, United States
We performed cross-species comparison to determine how the human default mode network (DMN) connectivity pattern compares to non-hominid primates. We characterized and compared the resting-state network functional organisation in humans, macaques, marmosets, and mouse lemurs using functional and anatomical atlases. We found decreased engagement of mPFC (medial prefrontal cortex) in all non-hominid primates “DMN-like” compared to humans. Another network involving mPFC was identified in all non-hominid primates but not in humans. Altogether, our results show that high order networks often assumed to be shared across primates diverge considerably between non-hominid species and humans.
|
|
 |
0574.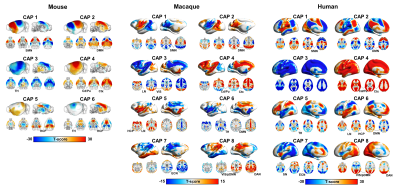 |
Evolutionarily conserved fMRI network dynamics in the human, macaque and mouse brain
Daniel Gutierrez-Barragan1, Stefano Panzeri2, Ting Xu3, and Alessandro Gozzi1
1Functional Neuroimaging Laboratory,, Istituto Italiano di Tecnologia, CNCS, Rovereto, Italy, 2Neural Computation Laboratory, Istituto Italiano di Tecnologia, CNCS, Rovereto, Italy, 3Center for the Developing Brain, Child Mind Institute, New York, NY, United States
Recent work revealed that resting-state fMRI (rsfMRI) network dynamics in the mouse brain governed by infraslow transitions between a limited set of recurring BOLD co-activation patterns. Here we extend these findings to the macaque and awake human brain, showing that in both these higher mammalian species rsfMRI timeseries can be similarly deconstructed into a set of oscillatory coactivation patterns, whose occurrence is phase-locked to intrinsic global fMRI Signal (GS) fluctuations. Our results reveal a fundamental, evolutionarily conserved spatiotemporal structure of resting-state fMRI activity.
|
|
 |
0575.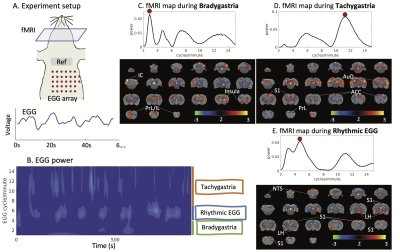 |
Power and Frequency Fluctuations of Gastric Electrical Activity Modulate fMRI Activity in Rat Brains
Jiayue (Cherry) Cao1, Xiaokai Wang1, Yizhen Zhang2, and Zhongming Liu1,2
1Biomedical Engineering, University of Michigan, ANN ARBOR, MI, United States, 2Electrical Engineering and Computer Science, University of Michigan, ANN ARBOR, MI, United States
The stomach-brain interaction is critical for regulating gastric function. Prior studies suggest that the brain maintains a slow rhythm coupled to the gastric slow wave – a rhythmic activity that paces the stomach. Here, we find a brain network encodes the frequency and power fluctuations of gastric rhythms in the resting state. Rhythmic activity of the stomach primarily engages the NTS-insula-somatosensory network. Dysrhythmic activities, such as bradygastria and tachygastria engage additional regions in the anterior cingulate cortex, prelimbic cortex, and infralimbic cortex. The alternation of dysrhythmic and pace-making activity in the stomach causes activity fluctuations across a central gastric network.
|
|
0576.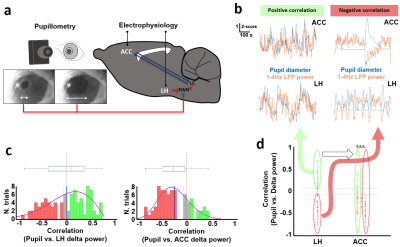 |
Identifying functional correlations between lateral hypothalamus and cingulate cortex underlying brain state-dependent pupil dynamics
Kengo Takahashi1,2, Filip Sobczak1,2, Patricia Pais-Roldán3, and Xin Yu1,4
1High-field Magnetic Resonance, Max Planck Institute for Biological Cybernetics, Tübingen, Germany, 2Graduate Training Centre of Neuroscience, Tübingen, Germany, 3Institute of Neuroscience and Medicine (INM-4), Forschungszentrum Jülich, Jülich, Germany, 4Athinoula A. Martinos Center, Massachusetts General Hospital and Harvard Medical School, Boston, MA, United States
The lateral hypothalamus (LH) is known to mediate different vigilance states and modulate pupil dilation through various neural populations. However, measuring subcortical neuronal activity non-invasively while assessing the brain state has remained challenging. Recently, it has been shown that the coupling between fMRI brain signals and pupil size fluctuations depends on the underlying brain state. In this work, we suggest that the synchronization of LH fMRI signals with pupil fluctuations may indicate modulation of the vigilance level of the brain.
|
||
0577.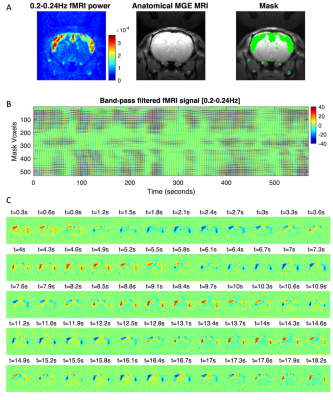 |
Resonant oscillatory modes in rat cortical activity revealed by ultra-fast fMRI
Joana Cabral1,2, Francisca F. Fernandes1, and Noam Shemesh1
1Champalimaud Research, Champalimaud Centre for the Unknown, Lisbon, Portugal, 2Life and Health Sciences Research Institute, University of Minho, Braga, Portugal
We provide evidence of the oscillatory nature of spontaneous BOLD signal fluctuations by performing a spectral analysis of fMRI signals recorded at unprecedented time resolution (TR = 38 ms) from a frontal brain slice of sedated and anesthetized rats. Oscillations peaking between 0.03 Hz and 0.25 Hz are detected in the cortex under sedation, drastically decreasing in power with deep anaesthesia. Notably, the power of BOLD signal oscillations is modulated in space and time, synchronizing in phase across distant bilateral brain areas, providing insights into the mechanisms underlying resting-state functional connectivity.
|
||
 |
0578.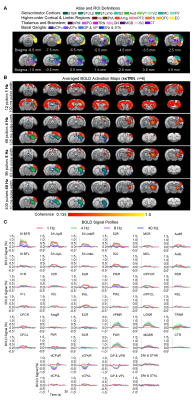 |
Inhibitory Thalamic Reticular Nucleus Drives Frequency Specific Brain-wide Responses
Xunda Wang1,2, Alex T. L. Leong1,2, Eddie C. Wong1,2, Teng Ma1,2,3, Pit Shan Chong4, Chi Him Poon4, Pek-Lan Khong3, Lee-Wei Lim4, and Ed X. Wu1,2
1Laboratory of Biomedical Imaging and Signal Processing, The University of Hong Kong, Hong Kong SAR, China, 2Department of Electrical and Electronic Engineering, The University of Hong Kong, Hong Kong SAR, China, 3Department of Diagnostic Radiology, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China, 4School of Biomedical Sciences, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Hong Kong SAR, China
Thalamic inhibition from thalamic reticular nucleus has been shown to provide critical gating upon thalamo-cortical interactions and exert selective modulation on information processing according to behavioral demands. However, where and how thalamic reticular nucleus exerts control of brain-wide neural activities over different spatial and temporal scales remained unclear. In this study, we demonstrate for the first time the frequency specific brain-wide responses driven by inhibitory somatosensory thalamic reticular nucleus using optogenetic fMRI. Such frequency specific engagements of brain-wide neural activities could underlie selective modulation of local circuits versus global networks in different brain functions.
|
The International Society for Magnetic Resonance in Medicine is accredited by the Accreditation Council for Continuing Medical Education to provide continuing medical education for physicians.