1Department of Biomedical Imaging and Radiological Sciences, National Yang-Ming University, Taipei, Taiwan, Taipei, Taiwan, 2Department of Neurosurgery, Neurological Institute, Taipei Veteran General Hospital, Taipei, Taiwan, Taipei, Taiwan, 3School of Medicine, National Yang-Ming University, Taipei, Taiwan, Taipei, Taiwan, 4Brain Research Center, National Yang-Ming University, Taipei, Taiwan, Taipei, Taiwan, 5Department of Radiology, Taipei Veteran General Hospital, Taipei, Taiwan, Taipei, Taiwan, 6Department of Medical Imaging, Cheng-Hsin General Hospital, Taipei, Taiwan, Taipei, Taiwan, 7Molecular and Genetic Imaging Core, Taiwan Animal Consortium, Taipei, Taiwan, Taipei, Taiwan, 8Institute of Biophotonics, National Yang-Ming University, Taipei, Taiwan, Taipei, Taiwan
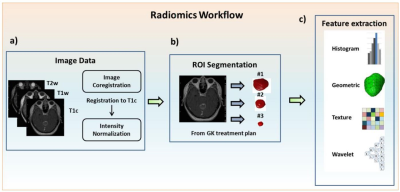
Figure 1. Image processing steps and radiomics flowchart
(a) Image acquisition of T1cwc, T1w and T2w images, registration of T1w and T2w to T1c images and intensity normalization. (b) ROI delineation for the treatment planning of GKRS by experienced neurosurgeons and radiologists on all MRI slices covering tumor regions. (c) Extraction of radiomic features from the tumor ROIs on the MRIs.
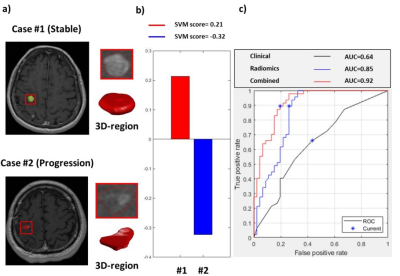
Figure 2. Representative cases and the model performance in predicting local tumor control
(a) T1c images and reconstructed 3D tumor models. (b) The SVM scores in two representative cases based on the combination of radiomic and clinical features. (c) Receiver operating characteristic curves and the area under the curves (AUCs) of three SVM classification models.