Dipika Sikka1,2, Nanyan Zhu3, Chen Liu4, Scott Small5, and Jia Guo6
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2VantAI, New York, NY, United States, 3Department of Biological Sciences and the Taub Institute, Columbia University, New York, NY, United States, 4Department of Electrical Engineering and the Taub Institute, Columbia University, New York, NY, United States, 5Department of Neurology, the Taub Institute, the Sergievsky Center, Radiology and Psychiatry, Columbia University, New York, NY, United States, 6Department of Psychiatry, Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States
1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2VantAI, New York, NY, United States, 3Department of Biological Sciences and the Taub Institute, Columbia University, New York, NY, United States, 4Department of Electrical Engineering and the Taub Institute, Columbia University, New York, NY, United States, 5Department of Neurology, the Taub Institute, the Sergievsky Center, Radiology and Psychiatry, Columbia University, New York, NY, United States, 6Department of Psychiatry, Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States
Contrast produced by a deep learning algorithm shows comparable predictions of gadolinium uptake for brain and breast lesion enhancement, using a single T1-weighted pre-contrast scan.
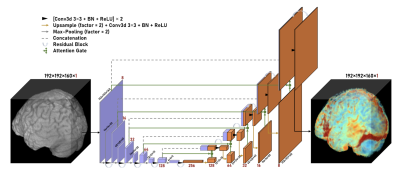
Figure 1. 3D Residual Attention U-Net architecture for brain lesion data. The network consists of 6 encoding layers (purple) and 6 decoding layers (orange). Spatial dimension decreases by 2 and channel dimension increases by 2 as the data propagates through the encoding layers while the reverse happens along the decoding layers. A full scan is then returned as the model output, as the prediction of the entire scan.
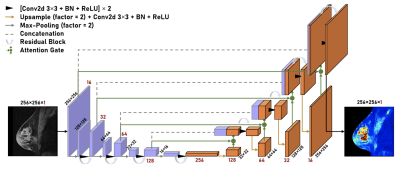
Figure 2. 2D Residual Attention U-Net architecture for breast lesion data. The network consists of 5 encoding layers (purple) and 5 decoding layers (orange). Spatial dimension decreases by 2 and channel dimension increases by 2 as the data propagates through the encoding layers while the reverse happens along the decoding layers. A single slice of the predicted scan is then returned as the model output.
