Zepeng Wang1,2, Yahang Li1,2, and Fan Lam1,2
1Department of Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, Urbana, IL, United States
1Department of Bioengineering, University of Illinois Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, Urbana, IL, United States
We presented further optimized J-resolved MRSI for high-resolution, 3D metabolite, and neurotransmitter mapping. Estimation-theoretic TE selection within a union-of-subspaces framework was analyzed. Simultaneously mapping of major metabolites, Glx, and GABA are provided.
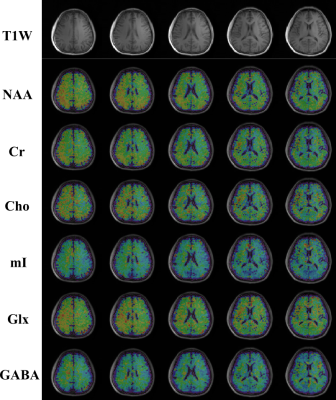
Figure 5. High-resolution and high-SNR metabolite and neurotransmitter maps estimated from the in vivo data (a 3.4×3.4×5.3mm3 nominal resolution). Anatomical images (T1w) across different slices from the 3D imaging volume are shown in the top row, and maps of different metabolites, as well as the Glx and GABA components, are shown in subsequent rows.
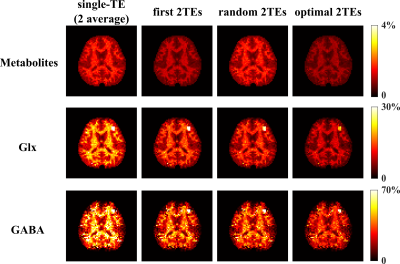
Figure 2. Monte-Carlo analysis showing the normalized standard deviation (std) of the coefficient estimates for different components from the UoSS fitting (Row 1: metabolite, Row 2: Glx, and Row 3: GABA). Several alternative TE choices with an equivalent acquisition time are considered for a 2-TE case. Specifically, columns 1-4 display the std maps for the case of single-TE (35ms, 2 average), first 2 TEs (35 and 50ms), random 2 TEs (50 and 110ms), and optimized 2 TEs (65 and 80ms), respectively, The best std maps were achieved by the optimized 2 TEs, consistent with the CRLB prediction.
