Weimin Zhang1, Chenyu He1, Xiaojun Guan2, Xiaojun Xu2, Hongjiang Wei3, and Yuyao Zhang1,4,5
1School of Information Science and Technology, ShanghaiTech University, Shanghai, China, 2Department of Radiology, The Second Affiliated Hospital, Zhejiang, China, 3Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai, China, 4Shanghai Engineering Research Center of Intelligent Vision and Imaging, ShanghaiTech University, Shanghai, China, 5iHuman Institute, ShanghaiTech University, Shanghai, China
1School of Information Science and Technology, ShanghaiTech University, Shanghai, China, 2Department of Radiology, The Second Affiliated Hospital, Zhejiang, China, 3Institute for Medical Imaging Technology, School of Biomedical Engineering, Shanghai, China, 4Shanghai Engineering Research Center of Intelligent Vision and Imaging, ShanghaiTech University, Shanghai, China, 5iHuman Institute, ShanghaiTech University, Shanghai, China
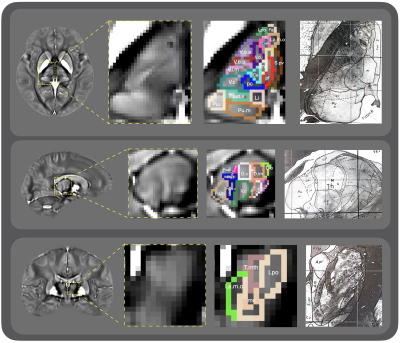
we present a multi-atlas label fusion framework to delineate a total number of 64 thalamic nuclei using both 7T and 3T QSM images. By combining group-wise registration, label propagation, and label fusion processes, we achieve a 64 thalamic nuclei parcellation map in 3T QSM atlas space.
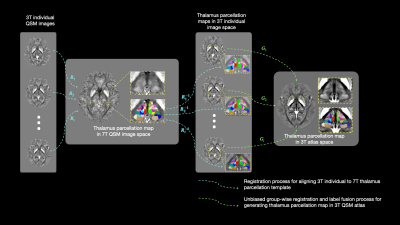
Fig. 1. Flowchart of multi-atlas label fusion framework. Firstly, a non-linear registration is performed to align 3T individual to 7T QSM image space; then inverted deformation fields are applied onto 7T thalamic sub-nucleus parcellation map to generate individual estimation maps; finally, joint label fusion method fuses these parcellation map into 3T atlas space.