Bonian Lu1, Rangaprakash Deshpande2, Madhura Ingalhalikar3, and Gopikrishna Deshpande1
1Electrical and Computer Engineering, AU MRI Research Center, Auburn University, Auburn, AL, United States, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Medical School and Harvard-MIT Health Sciences and Technology, Charlestown, MA, United States, 3Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India
1Electrical and Computer Engineering, AU MRI Research Center, Auburn University, Auburn, AL, United States, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Medical School and Harvard-MIT Health Sciences and Technology, Charlestown, MA, United States, 3Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India
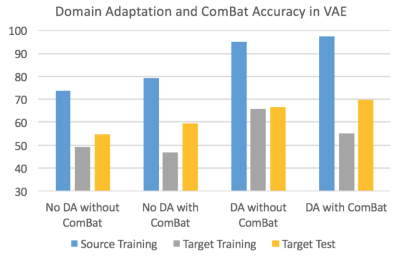
We applied deep learning-based domain adaptation and statistical ComBat harmonization for improving diagnostic accuracy from multi-site neuroimaging data and show robust performance of our approach in the context of diagnostic classification of ASD.
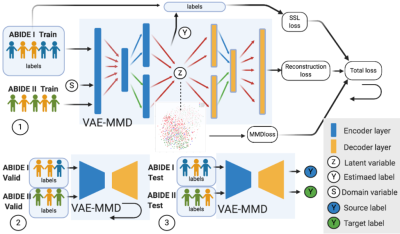
Figure 1. Three major steps in learning VAE-MMD model. 1. For training we input both ABIDE I with labels and ABIDE II training datasets into VAE-MMD model. The original t-SNE figure and domain adapted t-SNE figure (both shown in Fig.2) are generated in the beginning and last iteration of this process. The total loss is constructed by SSL loss, reconstruction loss and MMD loss. 2.For validation, ABIDE I and ABIDE II validation dataset are used to fine-tuning the hyperparameters α and β. 3. For testing, ABIDE I & ABIDE II test dataset are used in testing model to evaluate the model’s performance