Maximilian Pietsch1,2, Daan Christiaens2,3, Jana Hutter2,4, Lucilio Cordero-Grande2, Anthony N. Price2,4, Emer Hughes2, A. David Edwards2, Joseph V. Hajnal2,4, Serena J. Counsell2, Jonathan O'Muircheartaigh1,2,5,6, and J-Donald Tournier2,4
1Forensic & Neurodevelopmental Sciences, King's College London, London, United Kingdom, 2Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King’s College London, Kings Health Partners, St. Thomas Hospital, London, SE1 7EH, UK, King's College London, London, United Kingdom, 3Department of Electrical Engineering (ESAT/PSI), KU Leuven, Leuven, Belgium, 4Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 5Department of Perinatal Imaging & Health, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 6MRC Centre for Neurodevelopmental Disorders, King's College London, London, United Kingdom
1Forensic & Neurodevelopmental Sciences, King's College London, London, United Kingdom, 2Centre for the Developing Brain, School of Biomedical Engineering and Imaging Sciences, King’s College London, Kings Health Partners, St. Thomas Hospital, London, SE1 7EH, UK, King's College London, London, United Kingdom, 3Department of Electrical Engineering (ESAT/PSI), KU Leuven, Leuven, Belgium, 4Biomedical Engineering Department, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 5Department of Perinatal Imaging & Health, School of Biomedical Engineering and Imaging Sciences, King's College London, London, United Kingdom, 6MRC Centre for Neurodevelopmental Disorders, King's College London, London, United Kingdom
We decompose neonatal HARDI signal into four components that together capture the orientation and spatio-temporal dependency of signal in neonatal white matter, grey matter and CSF.
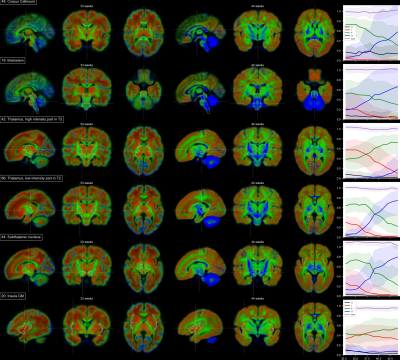
Outlines of anatomical structures defined in the anatomical atlas 10 overlaid onto the three diffusion-derived contrasts I (red), F (green) and S (blue). From left to right: compartment volume fraction maps for neonates at 33 weeks and 44 weeks and the mean and 90% IQR of the voxel-wise component volume fractions in the atlas over gestational age.
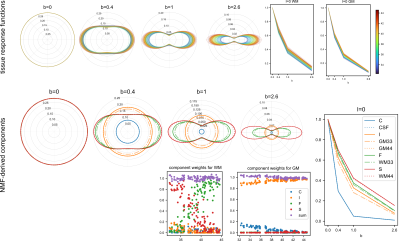
Top: angular profile of the WM response functions normalised to the b=0, l=0 signal and b-value dependency of the normalised WM and GM response functions for all subjects, colour-coded by gestational age in weeks. Bottom: The four-component sNMF model components and the weights associated with the sampled WM and GM response functions plotted against age for all subjects in the cohort. Note that no temporal constraints were used in the model.
