Yang Zhou1, Peter van Zijl2,3, Chongxue Bie2,3,4, Jiadi Xu2,3, Xin Liu1, and Nirbhay N. Yadav2,3
1Institute of Biomedical and Health Engineering, Shenzhen Institutes of Advanced Technology, Shenzhen, China, 2The Russell H. Morgan Department of Radiology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 4Department of Information Science and Technology, Northwest University, Xian, China
1Institute of Biomedical and Health Engineering, Shenzhen Institutes of Advanced Technology, Shenzhen, China, 2The Russell H. Morgan Department of Radiology, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 4Department of Information Science and Technology, Northwest University, Xian, China
We show that electrostatic binding interactions between substrates and a macromolecular matrix can be detected using IMMOBILISE MRI. A model is developed to quantitatively describe relayed NOE based magnetization transfer during such ionic bonding.
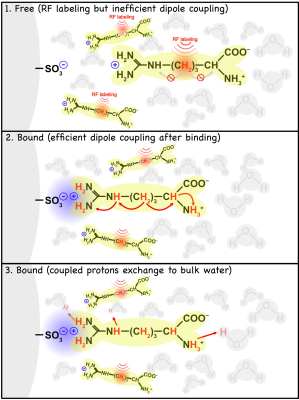
Figure 1. Principles of rNOE saturation transfer during electrostatic binding of a small molecule to an immobile medium with charged groups (here -SO3- ). The aliphatic protons of free arginine are efficiently labelled by a radio-frequency (RF) pulse, and there is no rNOE based transfer within the molecule, which is in fast-tumbling mode. Bound arginine molecules are in the slow-tumbling mode, where there is efficient intramolecular rNOE transfer from aliphatic protons to exchangeable protons, and finally to water via chemical exchange processes.
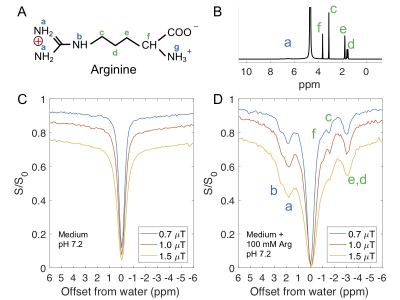
Figure 2. CEST MRI of arginine solutions mixed with ion-exchange medium (with functional group -SO3-). (A) The chemical structure of arginine; (B) 1D 1H NMR of arginine (5% H2O/95% D2O, pD 7.2), peak assignments are based on previous studies.6,7 (C-D) Z-spectra of ion-exchange resin medium alone (C), medium with arginine (100 mM, pH of 7.2) (D).