Xiaokai Wang1, Jiayue Cao1, Minkyu Choi2, and Zhongming Liu3
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 2Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States, 3Biomedical Engineering, Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States
1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 2Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States, 3Biomedical Engineering, Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States
A deep learning-based pipeline for fully automatic analysis of gastrointestinal MRI was established to characterize gastric motility. Results from this analysis are consistent with EGG measurements, and in addition, shed light on more detailed spatial characteristics of gastric motility.
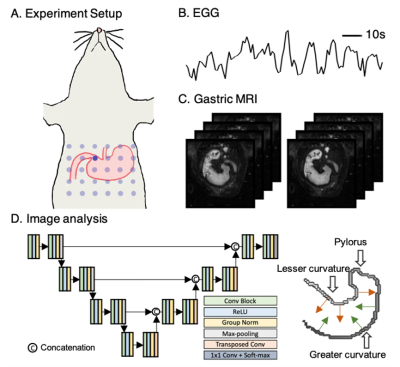
Fig. 1 Simultaneous MRI acquisition and EGG recording helped in validating our newly proposed deep learning-based fully automatic GI MR image analysis pipeline. (A) layouts the scheme for the 30-channel electrode array attached over the skin surface on top of the stomach. (B) and (C) show examples of pre-processed EGG signals and acquired GI MR images, respectively. (D) displays the 2D U-net for segmenting stomach from MR images and the extraction scheme of peristaltic displacements perpendicular to the gastric long axis along the greater and lesser curvatures.
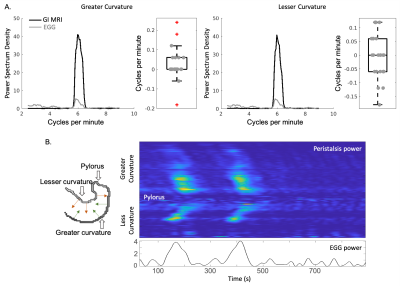
Fig. 2 The frequency and power of the estimated displacements along the greater and lesser curvatures were validated against the frequency and power of EGG, respectively. (A) shows the frequency distribution of EGG in gray lines and dots and of estimated displacements along the greater and lesser curvatures in black lines and boxes. We then targeted on 6.25 CPM, (B) shows the time-evolving power of the estimated peristalsis within the narrow frequency band along the greater and lesser curvatures in the upper panel and of EGG shown within the same narrow band in the lower panel.