Hwihun Jeong1, Hyeong-Geol Shin1, Sooyeon Ji1, Jinhee Jang2, Hyun-Soo Lee3, Yoonho Nam4, and Jongho Lee1
1Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 2Department of Radiology, Seoul St Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea, Republic of, 3Siemens healthineers Ltd, Seoul, Korea, Republic of, 4Division of Biomedical Engineering, Hankuk University of Foreign Studies, Yongin, Korea, Republic of
1Department of Electrical and Computer Engineering, Seoul National University, Seoul, Korea, Republic of, 2Department of Radiology, Seoul St Mary’s Hospital, College of Medicine, The Catholic University of Korea, Seoul, Korea, Republic of, 3Siemens healthineers Ltd, Seoul, Korea, Republic of, 4Division of Biomedical Engineering, Hankuk University of Foreign Studies, Yongin, Korea, Republic of
We
developed a deep learning-based T2 mapping method with retrospective
B1+ estimation for a double-echo TSE sequence and applied it to χ-separation
and an MS patient.
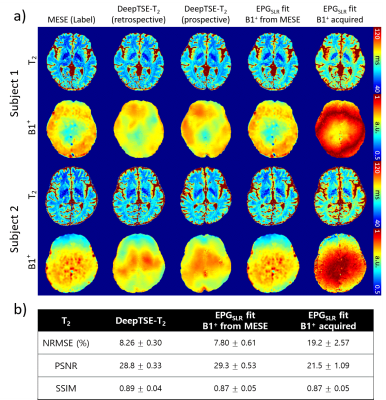
Figure 2. (a) Results of DeepTSE-T2 and conventional EPGSLR fitting with
extra B1+ information. B1+ maps in the fourth and fifth columns
are used for the EPGSLR fitting. In the acquired B1+ map,
values above 1 were flipped with respect to 1, assuming symmetry. (b) NRMSE,
PSNR, SSIM of the T2 maps with respect to the labels. DeepTSE-T2 provides a high-quality T2
map, comparable to those of the EPGSLR fitting with B1+
map from MESE.
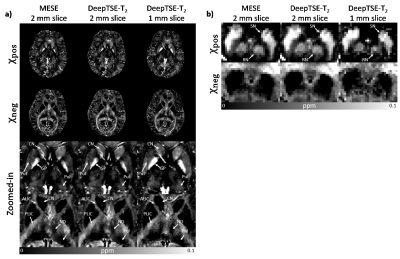
Figure 5. Results of χ-separation using DeepTSE-T2 map: ALIC -
anterior limb of internal capsule, CN - caudate nucleus, GP - globus pallidus,
ND - nucleus dorsomedialis, PLIC - Posterior limb of internal capsule, Pul -
pulvinar, Put - putamen, RN - red nucleus, and SN - substantia nigra. In the
case of 2 mm slice thickness, the positive and negative susceptibilities using DeepTSE-T2
show similar results with those using MESE. 1 mm slice χ-separation using DeepTSE-T2 is also
illustrated, demonstrating feasibility of high resolution χ
-separation.
