Yibo Zhao1,2, T. Kevin Hitchens3,4, Michele Herneisey5, Jelena M. Janjic5, Rong Guo1,2, Yudu Li1,2, and Zhi-Pei Liang1,2
1Department of Electrical and Computer Engineering, University of Illinois, Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois, Urbana-Champaign, Urbana, IL, United States, 3Animal Imaging Center, University of Pittsburgh, Pittsburgh, PA, United States, 4Department of Neurobiology, University of Pittsburgh, Pittsburgh, PA, United States, 5Graduate School of Pharmaceutical Sciences, Duquesne University, Pittsburgh, PA, United States
1Department of Electrical and Computer Engineering, University of Illinois, Urbana-Champaign, Urbana, IL, United States, 2Beckman Institute for Advanced Science and Technology, University of Illinois, Urbana-Champaign, Urbana, IL, United States, 3Animal Imaging Center, University of Pittsburgh, Pittsburgh, PA, United States, 4Department of Neurobiology, University of Pittsburgh, Pittsburgh, PA, United States, 5Graduate School of Pharmaceutical Sciences, Duquesne University, Pittsburgh, PA, United States
A
novel method is proposed for accelerated high-resolution 19F-MRSI of multiple perfluorocarbon nanoemulsions, using EPSI trajectories and
union-of-subspaces modeling with pre-learned subspaces. The proposed method has
been validated using both simulation and experimental data.
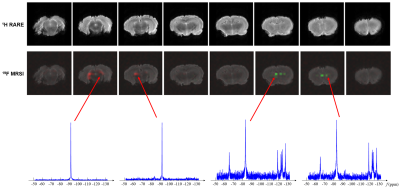
Figure 4. Results obtained from a fixed
rat brain injected with PCE and PFOB NEs. PCE (red) and PFOB (green) maps are overlaid
on the 1H anatomical image, and
representative spectra are shown. As can be seen, the proposed method obtained
PCE and PFOB maps and spectra as expected.
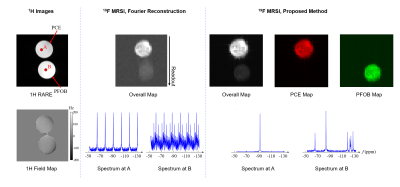
Figure 3. Proton structural image, field
map, and comparison of Fourier-based and proposed 19F-MRSI processing results obtained from a
phantom. As can be seen, Fourier-based processing results suffered from spatial
chemical shift artifacts and spectral aliasing artifacts, while the proposed
method produced artifact-free spatiospectral functions for PCE and PFOB.
