Suphachart Leewiwatwong1, Junlan Lu2, David Mummy3, Isabelle Dummer3,4, Kevin Yarnall5, Ziyi Wang1, and Bastiaan Driehuys1,2,3
1Biomedical Engineering, Duke University, Durham, NC, United States, 2Medical Physics, Duke University, Durham, NC, United States, 3Radiology, Duke University, Durham, NC, United States, 4Bioengineering, McGill University, Montréal, QC, Canada, 5Mechanical Engineering and Materials Science, Duke University, Durham, NC, United States
1Biomedical Engineering, Duke University, Durham, NC, United States, 2Medical Physics, Duke University, Durham, NC, United States, 3Radiology, Duke University, Durham, NC, United States, 4Bioengineering, McGill University, Montréal, QC, Canada, 5Mechanical Engineering and Materials Science, Duke University, Durham, NC, United States
We demonstrate
a 3D convolutional neural network that
facilitates quantitative image analysis by providing a thoracic cavity
segmentation using only the 129Xe MRI. The CNN achieves a Dice score of 0.955 vs. the gold standard
expert reader segmentation.
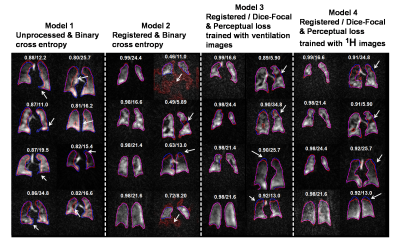
Figure 4. Segmentations from different models.
For each model, the 4 best (left) and worst (right) segmentations are shown
with Dice score/SNR (these were calculated from the whole 3D volume). The models’
segmentations are shown in red, those from the expert readers in blue, and
their overlaps in purple. White arrows indicate areas where models deviated
from ground truth. From this comparison, the model trained with registered data
performs better than unprocessed data but struggles with low-SNR and poor
ventilation images. Our proposed method (model 3 & 4) helps solve this
problem.
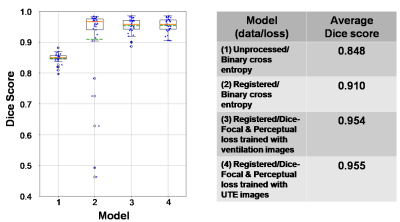
Figure 5. Box plot and table of Dice score for each model tested with the “pristine”
test dataset. The orange lines indicate the median of the score while the dash
green lines indicate the mean. By comparing model 1 and 2, the plot shows the
improvement in using the registered dataset for training 3D-CNN but with a
drawback shown by low-score outliers due to poor ventilation and low-SNR images. Our
proposed method (model 3 & 4) solves the problem and eliminates these
outliers.