Noemi G. Gyori1,2, Marco Palombo1, Christopher A. Clark2, Hui Zhang1, and Daniel C. Alexander1
1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Great Ormond Street Institute of Child Health, University College London, London, United Kingdom
1Centre for Medical Image Computing, University College London, London, United Kingdom, 2Great Ormond Street Institute of Child Health, University College London, London, United Kingdom
We demonstrate that training on the observed parameter-combination distribution may be advantageous for detecting small variations in healthy tissue, whereas for detecting atypical tissue abnormalities sampling from a uniform training data distribution may be favourable.
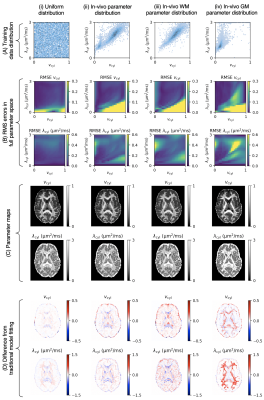
Figure 2. Panel (A): Different training data distribution strategies: (i) uniform distribution, (ii) in-vivo parameter combinations from traditional model fitting, (iii) in-vivo white matter parameter combinations and (iv) in-vivo grey matter parameter combinations. Panel (B): RMS errors of vcyl estimates (top) and λcyl estimates (bottom) at different parameter combinations. Panel (C): Parameter maps estimated from each training data distribution. Panel (D): Difference between parameter maps in panel (C) and parameter maps from traditional model fitting in Figure 1.
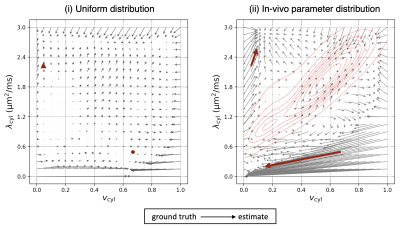
Figure 4. The bias in parameter estimates for (i) the uniform training data distribution and for (ii) the in-vivo parameter distribution. The arrows start from the ground truth of each parameter combination and end at the estimates. Red arrows mark the tissue abnormalities shown in Figure 3. In (ii), the red contours show the training data density. Multiple regions of the parameter space act as “sinks” towards which estimates of nearby parameters are biased.
