João Pedro de Almeida Martins1,2, Markus Nilsson1, Björn Lampinen3, Marco Palombo4, Carl-Fredrik Westin5,6, and Filip Szczepankiewicz1,5,6
1Department of Clinical Sciences, Radiology, Lund University, Lund, Sweden, 2Department of Radiology and Nuclear Medicine, St. Olav's University Hospital, Trondheim, Norway, 3Department of Clinical Sciences, Medical Radiation Physics, Lund University, Lund, Sweden, 4Centre for Medical Image Computing and Dept of Computer Science, University College London, London, United Kingdom, 5Radiology, Brigham and Women’s Hospital, Boston, MA, United States, 6Harvard Medical School, Boston, MA, United States
1Department of Clinical Sciences, Radiology, Lund University, Lund, Sweden, 2Department of Radiology and Nuclear Medicine, St. Olav's University Hospital, Trondheim, Norway, 3Department of Clinical Sciences, Medical Radiation Physics, Lund University, Lund, Sweden, 4Centre for Medical Image Computing and Dept of Computer Science, University College London, London, United Kingdom, 5Radiology, Brigham and Women’s Hospital, Boston, MA, United States, 6Harvard Medical School, Boston, MA, United States
We found that neural networks can vastly accelerate the fitting of high-dimensional microstructural models but cannot by themselves reduce the requirements on the acquisition protocol or correct for the degeneracy of the parameter estimation problem.
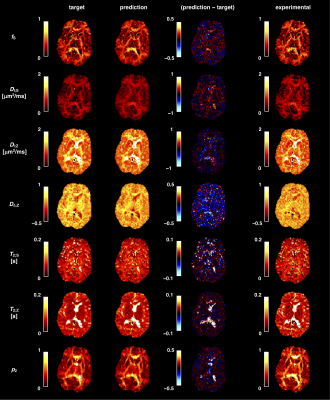
Figure 2. Deploying trained networks on previously unseen synthetic and in vivo data provides anatomically plausible parameter maps in under 10 s. The parameter maps corresponding to the synthetic dataset (second column) are compared to the corresponding ground-truth labels (first column), with difference maps being shown in the third column. Parameter maps obtained from applying a trained network on in vivo brain data are displayed in the fourth column.
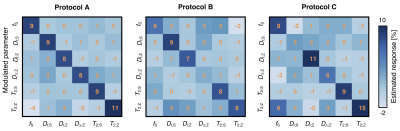
Figure 5. Sensitivity of different acquisition protocols to 10% parameter modulations. The matrices display the relation between an induced parameter change and the observed response. When a single parameter on the y-axis is modulated by 10%, the response can be read in all other parameters along the x-axis. An ideal network would report a diagonal matrix with the value 10% on the diagonal, and zero otherwise. Protocol A appears sensitive in all parameters, whereas Protocols B and C lack sensitivity to DΔ;Z and DI;S, respectively.