Manuel A. Morales1,2, Maaike van den Boomen2,3,4, Christopher Nguyen2,4, Jayashree Kalpathy-Cramer2, Bruce R. Rosen1,2, Collin Stultz 1,5,6, David Izquierdo-Garcia1,2, and Ciprian Catana2
1Health Sciences & Technology, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Radiology, Athinoula A. Martinos Center for Biomedical Imaging, MGH, HMS, Charlestown, MA, United States, 3Radiology, University Medical Center Groningen, Groningen, Netherlands, 4Cardiovascular Research Center, MGH, HMS, Charlestown, MA, United States, 5Cardiology, Massachusetts General Hospital, Boston, MA, United States, 6Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States
1Health Sciences & Technology, Massachusetts Institute of Technology, Cambridge, MA, United States, 2Radiology, Athinoula A. Martinos Center for Biomedical Imaging, MGH, HMS, Charlestown, MA, United States, 3Radiology, University Medical Center Groningen, Groningen, Netherlands, 4Cardiovascular Research Center, MGH, HMS, Charlestown, MA, United States, 5Cardiology, Massachusetts General Hospital, Boston, MA, United States, 6Electrical Engineering and Computer Science, MIT, Cambridge, MA, United States
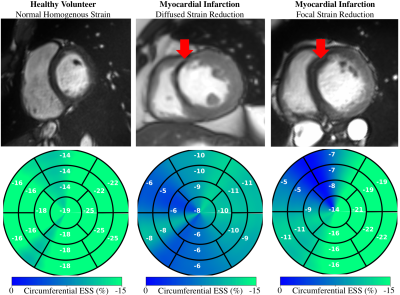
We developed and validated against tagging-MRI, an automated workflow for global and regional myocardial strain analysis to quantitatively characterize cardiac mechanics based on ubiquitously acquired cine-MRI data. Applications in patients showed global and asymmetric abnormalities.
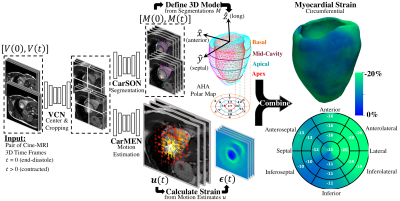
Overview of proposed workflow. VCN centers and crops the input pair of cine-MRI frames. Tissue labels generated by CarSON are used to build an anatomical model. Motion estimates derived from CarMEN are used to calculate strain measures, and these estimates are combined with the anatomical model to enable global and regional strain analyses.