Till Huelnhagen1,2,3, Omar Al Louzi4, Mário João Fartaria1,2,3, Lynn Daboul4, Pietro Maggi5,6, Cristina Granziera7,8,9, Meritxell Bach Cuadra2,3,10, Jonas Richiardi2, Daniel S Reich4, Tobias Kober1,2,3, and Pascal Sati4,11
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3Signal Processing Laboratory (LTS 5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4Translational Neuroradiology Section, National Institute of Neurological Disorders and Stroke, National Institutes of Health (NIH), Bethesda, MD, United States, 5Department of Neurology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 6Cliniques universitaires Saint-Luc, Université catholique de Louvain, Brussels, Belgium, 7Neurologic Clinic and Policlinic, Departments of Medicine, Clinical Research and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 8Translational Imaging in Neurology (ThINk) Basel, Department of Medicine and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 9Research Center for Clinical Neuroimmunology and Neuroscience (RC2NB) Basel, University Hospital Basel and University of Basel, Basel, Switzerland, 10Medical Image Analysis Laboratory (MIAL), Centre d'Imagerie BioMédicale (CIBM), University of Lausanne, Lausanne, Switzerland, 11Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, United States
1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3Signal Processing Laboratory (LTS 5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 4Translational Neuroradiology Section, National Institute of Neurological Disorders and Stroke, National Institutes of Health (NIH), Bethesda, MD, United States, 5Department of Neurology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 6Cliniques universitaires Saint-Luc, Université catholique de Louvain, Brussels, Belgium, 7Neurologic Clinic and Policlinic, Departments of Medicine, Clinical Research and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 8Translational Imaging in Neurology (ThINk) Basel, Department of Medicine and Biomedical Engineering, University Hospital Basel and University of Basel, Basel, Switzerland, 9Research Center for Clinical Neuroimmunology and Neuroscience (RC2NB) Basel, University Hospital Basel and University of Basel, Basel, Switzerland, 10Medical Image Analysis Laboratory (MIAL), Centre d'Imagerie BioMédicale (CIBM), University of Lausanne, Lausanne, Switzerland, 11Department of Neurology, Cedars-Sinai Medical Center, Los Angeles, CA, United States
This work proposes an improved version of a convolutional neural
network for the automated assessment of the central vein sign in brain MRI that
can accurately classify lesions of all central vein sign types without
requiring manual preselection.
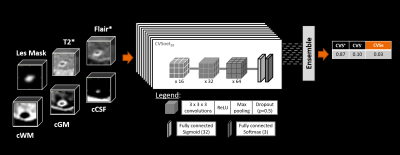
Figure 2: CNN architecture: An ensemble of 10 parallel
networks was used for classifying lesion patches as either CVS+, CVS−, or CVSe. With
regard to the previous CVSNet implementation, a third output class for CVSe was
added, and different combinations of input channels were tested to investigate
their impact on the classification performance.
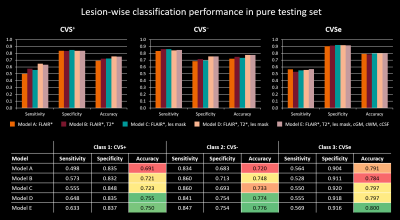
Figure 4: Lesion-wise performance comparison of the different
models in the pure testing set for each class. Overall performance was rather similar across the models but increased slightly with the number of used input
channels, except for model E, which showed comparable performance to model D
despite having three additional input channels. Accuracy levels are highlighted,
as this is regarded the most relevant metric with respect to a clinical
application. With accuracies of 75% to 80% in the best models, the network
performance is approaching levels of human inter-rater agreement.
