Thomas Lilieholm1, Ruiming Chen1, Ruvini Navaratna1, Daniel Seiter1, Walter F Block1,2,3, and Oliver Wieben1,2,3
1Medical Physics, University of Wisconsin at Madison, Madison, WI, United States, 2Biomedical Engineering, University of Wisconsin at Madison, Madison, WI, United States, 3Radiology, University of Wisconsin at Madison, Madison, WI, United States
1Medical Physics, University of Wisconsin at Madison, Madison, WI, United States, 2Biomedical Engineering, University of Wisconsin at Madison, Madison, WI, United States, 3Radiology, University of Wisconsin at Madison, Madison, WI, United States
ML-driven autonomous segmentation of placenta can be used in quantification of Zika virus infection biomarkers. Due to scarcity, we augmented data via geometric transform, finding rotation and reflection to yield improved segmentation models.
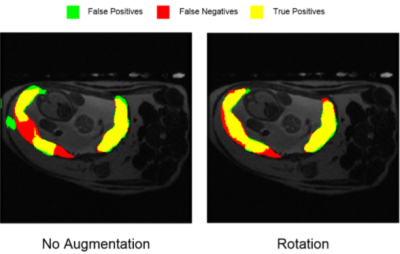
The segmentation produced for a single representative slice from the validation dataset, showing the results from training without any augmentation (left) versus training with augmentation via rotation. Comparison shows both a reduction in false positive, or improperly segmented pixels, and an increase in true positive, or properly-segmented pixels.
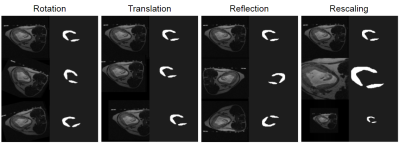
The geometric transformations applied to the original dataset to produce augmented data. Each original scan was subject to each type of transform twice, with different parameters. Rotation was up to ±30° about the physical z-axis. Translation was up to ±30 pixels along the physical x- or y-axes. Reflection was across either the physical x- or y-axes. Rescaling brought the placenta up to 500x500 pixels, or down to 150x150 pixels from the original 256x256 scan. Operations were performed using Matlab6.