Daniel Gutierrez-Barragan1, Stefano Panzeri2, Ting Xu3, and Alessandro Gozzi1
1Functional Neuroimaging Laboratory,, Istituto Italiano di Tecnologia, CNCS, Rovereto, Italy, 2Neural Computation Laboratory, Istituto Italiano di Tecnologia, CNCS, Rovereto, Italy, 3Center for the Developing Brain, Child Mind Institute, New York, NY, United States
1Functional Neuroimaging Laboratory,, Istituto Italiano di Tecnologia, CNCS, Rovereto, Italy, 2Neural Computation Laboratory, Istituto Italiano di Tecnologia, CNCS, Rovereto, Italy, 3Center for the Developing Brain, Child Mind Institute, New York, NY, United States
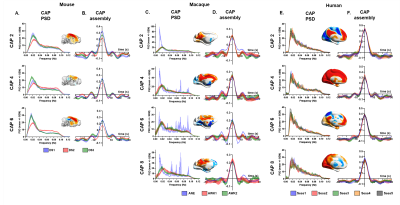
We present a set of dynamic features
governing whole-brain spontaneous fMRI activity in the mammalian brain (mouse, macaque
and human), delineating reproducible and recurrent BOLD co-activation topographies; state-dependent
dynamics; and evolutionary links between species.
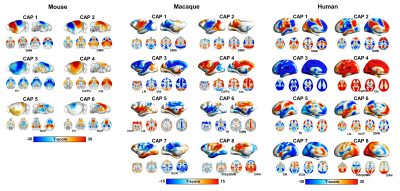
Fig2.Cross-species CAP topography
reveals evolutionarily conserved rsfMRI
network engagements. Co-activation
patterns (CAPs) from mice, macaques, and humans (p<0.05,Bonferroni
corrected). Red indicates significant co-activation, blue significant
co-deactivations. Abbreviations: Ctx-Cortex; SMN-Sensory-motor Network;
DMN-Default-Mode Network; TH-Thalamus; Cd/Pu-Caudate/Putamen;VIS-Visual
Network LN-Limbic Network;HCP-Hippocampus;
ECN-Executive-Control Network;DAN-Dorsal-Attention
Network; SN-Salience Network.