Kurt G Schilling1, Qi Yang2, Vishwesh Nath3, Rutger Fick4, Kristin P O'Grady1, Adam W Anderson2, Bennett A Landman1, and Seth A Smith1
1Vanderbilt University Medical Center, Nashville, TN, United States, 2Vanderbilt University, Nashville, TN, United States, 3NVIDIA, Bethesda, MD, United States, 4TheraPanacea, Paris, France
1Vanderbilt University Medical Center, Nashville, TN, United States, 2Vanderbilt University, Nashville, TN, United States, 3NVIDIA, Bethesda, MD, United States, 4TheraPanacea, Paris, France
We used extensive diffusion datasets to evaluate a large number of biophysical models (N=480) in the in vivo human spinal cord and found that certain compartments, and various constraints, are necessary to model diffusion in the cord and fully capture the changes in the diffusion MRI signal.
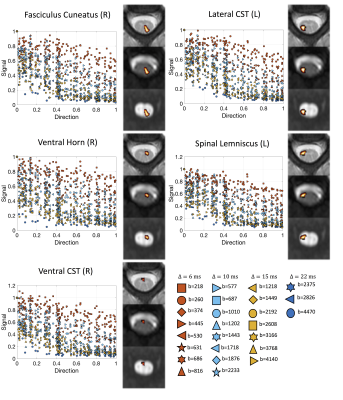
Figure 1. Data and regions of interest (ROIs). Data consisted of 2 subjects scanned with a 26-shell protocol (35 DWIs + 1 b0 per shell), resulting in 936 image volumes per subject. This covers a range of q-space, including multiple diffusion times, b-values, and angular directions. 16 ROIs were automatically extracted based on registration to the spinal cord atlas, and are shown overlaid on the structural image, the b0 image, and the mean diffusion weighted image.
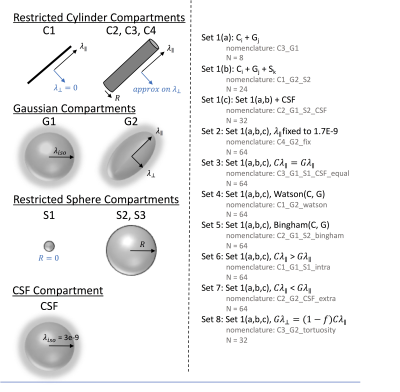
Figure 2. Multi-compartment models. Models consist of some combination of restricted cylinder compartments (C1-C4), Gaussian compartment (G1-G2), Restricted sphere compartments (S1-S3), and CSF compartments, with 8 different types of constraints used in the modeling process. This results in 480 different possible multi-compartment biophysical models of spinal cord microstructure.