Ashley Stewart1,2, Simon Daniel Robinson2,3,4, Kieran O'Brien1,2,5, Jin Jin1,2,5, Angela Walls6, Aswin Narayanan2, Markus Barth1,2,7, and Steffen Bollmann1,2,7
1Centre for Innovation in Biomedical Imaging Technology, University of Queensland, Brisbane, Australia, 2Centre for Advanced Imaging, University of Queensland, Brisbane, Australia, 3High Field MR Center, Department of Biomedical Imaging and Image-Guided Therapy, Medical University of Vienna, Vienna, Austria, 4Department of Neurology, Medical University of Graz, Graz, Austria, 5Siemens Healthcare Pty Ltd, Brisbane, Australia, 6Clinical & Research Imaging Centre, South Australian Health and Medical Research Institute, Adelaide, Australia, 7School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia
1Centre for Innovation in Biomedical Imaging Technology, University of Queensland, Brisbane, Australia, 2Centre for Advanced Imaging, University of Queensland, Brisbane, Australia, 3High Field MR Center, Department of Biomedical Imaging and Image-Guided Therapy, Medical University of Vienna, Vienna, Austria, 4Department of Neurology, Medical University of Graz, Graz, Austria, 5Siemens Healthcare Pty Ltd, Brisbane, Australia, 6Clinical & Research Imaging Centre, South Australian Health and Medical Research Institute, Adelaide, Australia, 7School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia
QSMxT provides a full QSM workflow including DICOM to BIDS conversion, robust masking strategies, phase unwrapping, background field correction, dipole inversion and region-of-interest analyses based on automated anatomical segmentations.
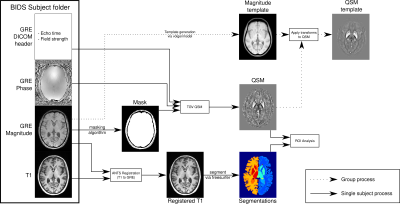
High-level Nipype data processing pipeline with visual illustrations of intermediate results and outputs. The inputs to single-subject processes are those of individual subjects, whereas group processes use the results from all subjects as input.
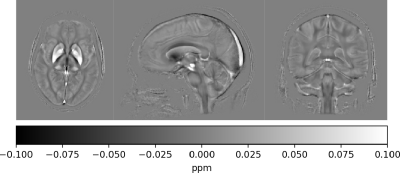
QSM template based on 45 subjects. Results were reconstructed using our QSMxT framework within our modular reconstruction and analysis ecosystem utilising the newly proposed masking technique free of anatomical assumptions. The template was generated using the minimum deformation averaging tool, volgenmodel.
