Brian J Soher1, Dinesh K Deelchand2, Sandeep Ganji3, Ralph Noeske4, Adam Berrington5, James Joers2, and Gulin Oz2
1Radiology, Duke University Medical Center, Durham, NC, United States, 2University of Minnesota, Minneapolis, MN, United States, 3Philips Healthcare, Rochester, MN, United States, 4GE Healthcare, Berlin, Germany, 5University of Nottingham, Nottingham, United Kingdom
1Radiology, Duke University Medical Center, Durham, NC, United States, 2University of Minnesota, Minneapolis, MN, United States, 3Philips Healthcare, Rochester, MN, United States, 4GE Healthcare, Berlin, Germany, 5University of Nottingham, Nottingham, United Kingdom
The
Vespa Inline Engine platform provides standardized data processing and
quantitative analysis of clinical single-voxel MRS data within the standard
DICOM workflow on GE, Philips and Siemens scanners.
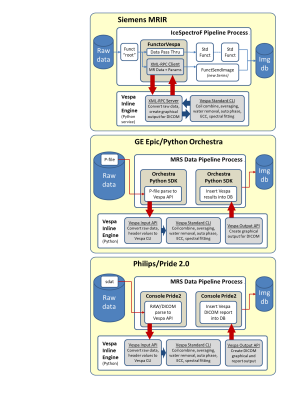
Figure 2. Manufacturer-specific
software implementations for Siemens (top), GE (middle) and Philips (bottom)
platforms. Standard manufacturer infrastructures (Siemens IDEA/ICE, GE Python
Orchestra and Philips PRIDE 2.0) are used to call the VIE module, which is a
pure Python implementation installed by copying a simple standard directory
structure onto a workstation. VIE results are sent to MR image databases as
DICOM ‘screenshot’ images of spectral plots and tabular metabolite fit results.
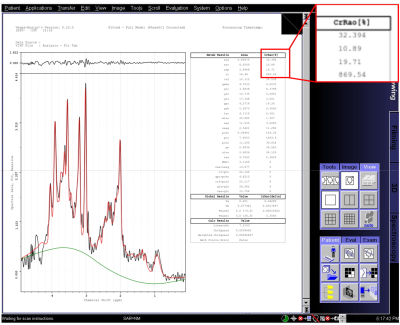
Figure
3. Fitting results for a test subject data taken on a Siemens Trio MR scanner.
Image resolution is 1024x1024 pixels. Spectral plots include raw data (black),
baseline estimate (green) and metabolite+macromolecule+baseline total fit (red),
metabolite fit values are in the table. Fitting time was approximately 90
seconds on the Siemens MRIR computer. An example of pixilation is shown in the
blown-up box, top right.
