Istvan N Huszar1, Karla L Miller1, and Mark Jenkinson1
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
A novel open-source registration
framework is released to automate the registration between sparsely sampled
(2-D) histology and 3-D post-mortem MRI data. Estimating also the through-plane
deformations of the sections yields submillimetre accuracy.
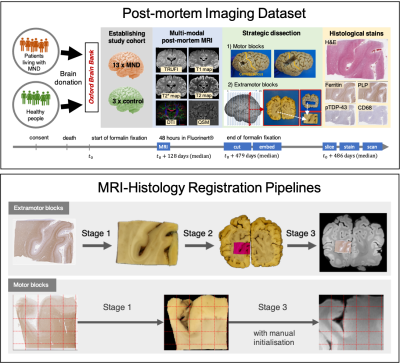
Figure 1. TOP: Acquisition
of post-mortem MRI, dissection photographs, and histology images. Motor and
extramotor blocks were sampled differently, yielding one or two photographic
intermediates (the block-face and the coronal slice images). BOTTOM: Histology-to-MRI
registration pipelines. Stage-specific transformations are combined to map the
high-resolution histology (0.5 mm/px) image into MRI space
(0.25 mm/vx). The MRI data is resampled by cubic spline interpolation to
produce a 2-D image.

