Andrew M Schmidt1, Arjun D Desai1, Lauren E Watkins2, Hollis Crowder3, Elka B Rubin1, Valentina Mazzoli1, Quin Lu4, Marianne Black1,3, Feliks Kogan1, Garry E Gold1,2, Brian A Hargreaves1,5, and Akshay S Chaudhari1,6
1Radiology, Stanford University, Stanford, CA, United States, 2Bioengineering, Stanford University, Stanford, CA, United States, 3Mechanical Engineering, Stanford University, Stanford, CA, United States, 4Philips Healthcare North America, Gainesville, FL, United States, 5Electrical Engineering, Stanford University, Stanford, CA, United States, 6Biomedical Data Science, Stanford University, Stanford, CA, United States
1Radiology, Stanford University, Stanford, CA, United States, 2Bioengineering, Stanford University, Stanford, CA, United States, 3Mechanical Engineering, Stanford University, Stanford, CA, United States, 4Philips Healthcare North America, Gainesville, FL, United States, 5Electrical Engineering, Stanford University, Stanford, CA, United States, 6Biomedical Data Science, Stanford University, Stanford, CA, United States
Manual-vs-automatic segmentation accuracy and T2 variations indicate that without model fine-tuning, deep-learning networks trained on a single dataset can generalize well to tissue relaxometry measurements but not exact morphology measurements, across subjects with varying health.
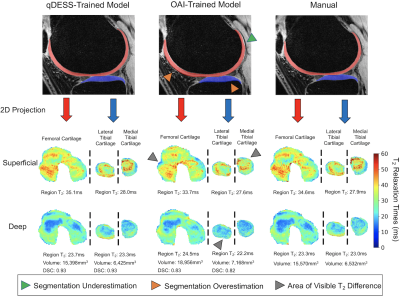
Comparison of manual and automatic segmentations from both models and respective 2D unrolled T2 maps in the right knee of a clinical patient in study 4. Also shown are the average T2 values from the superficial and deep cartilage regions, cartilage volumes, and DSC scores for the qDESS-trained and OAI-trained models. Arrows indicate examples of visually apparent differences in the automated segmentations and resultant T2 maps. These differences typically appear at the periphery of tissues, which have limited impact on subregion estimates.
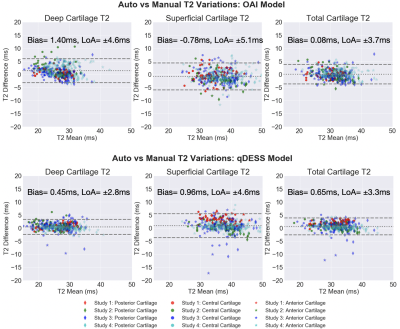
Bland-Altman plots for deep, superficial, and total femoral cartilage T2 relaxation times for both the OAI-trained and qDESS-trained models. Data is further stratified by study and anterior/central/posterior anatomic region. The T2 variations are minimal for both models and show no systematic error, however the limits of agreement for the qDESS-trained model for all cartilage layers are smaller.