Datta Singh Goolaub1,2 and Christopher Macgowan1,2
1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Translational Medicine, The Hospital for Sick Children, Toronto, ON, Canada
1Medical Biophysics, University of Toronto, Toronto, ON, Canada, 2Translational Medicine, The Hospital for Sick Children, Toronto, ON, Canada
A method to reduce clustering of
readout trajectories for CINE reconstructions is developed and demonstrated
through simulations. Improved image quality is presented.
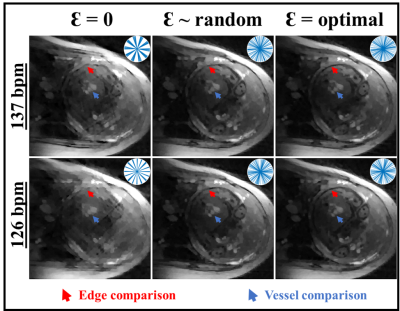
Representative reconstructions at HR
= 137 bpm and 126 bpm for trajectories without inter-block additional angles Ɛ (i.e. Ɛ = 0), random Ɛ (between 0 and 2π), and optimized Ɛ. The
trajectories in k-space are depicted on the top right corner of each
reconstruction. The arrows denote worst edge (blue) and vessel (red)
conspicuities with Ɛ = 0. Improvements are noticeable with random Ɛ. Further
edge improvement is observed with optimized Ɛ. bpm: beats per minute.
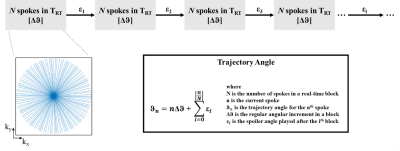
Framework
to reduce effects of clustering. After a block of N spokes spanning TRT, additional angles (εi) are played to reduce
coherence during cardiac gating. The k-space
from the first block corresponding to N = 64 spokes is shown with the blue
lines denoting the trajectories. Within each block the angular increment for
the trajectories is Δϑ allowing adequate k-space
coverage for reconstruction of real-time series.