Yamin Arefeen1, Borjan Gagoski2,3, and Elfar Adalsteinsson1,4,5
1Massachusetts Institute of Technology, Cambridge, MA, United States, 2Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 3Department of Radiology, Harvard Medical School, Boston, MA, United States, 4Harvard-MIT Health Sciences and Technology, Cambridge, MA, United States, 5Institute for Medical Engineering and Science, Cambridge, MA, United States
1Massachusetts Institute of Technology, Cambridge, MA, United States, 2Fetal-Neonatal Neuroimaging and Developmental Science Center, Boston Children’s Hospital, Boston, MA, United States, 3Department of Radiology, Harvard Medical School, Boston, MA, United States, 4Harvard-MIT Health Sciences and Technology, Cambridge, MA, United States, 5Institute for Medical Engineering and Science, Cambridge, MA, United States
A randomly undersampled, spiral-based trajectory yields 2-3x acceleration over fully sampled 3D-spiral acquisitions in 3D Magnetic Resonance Spectroscopy by inducing incoherent aliasing that can be resolved with SPICE-based low rank priors.
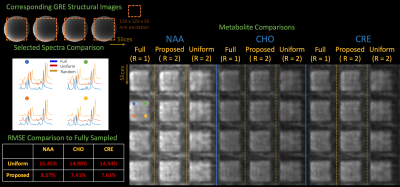
Figure 4: Metabolites maps, RMSE on image support, and example spectra from subspace reconstructions of fully sampled , uniformly undersampled (R = 2), and undersampled with the proposed trajectory (R = 2) on a spectroscopy phantom containing physiological concentrations of the major brain metabolites. The proposed trajectory yields similar spectra and metabolite maps to the fully sampled acquisition and achieves ~2x reduction in RMSE across the metabolites in comparison to uniform undersampling.
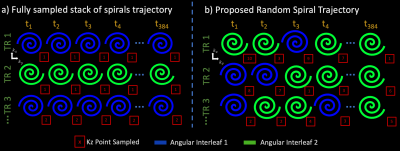
Figure 1: Schematic view of (a) the fully sampled 3D-spiral MRSI trajectory and (b) our proposed randomly undersampled trajectory. For demonstration purposes, we assume 1 temporal interleaf. In (a), the same angular interleaf and $$$k_z$$$ point is sampled at each timepoint in a TR. The proposed (b) trajectory samples random angular interleaves and $$$k_z$$$ points during each TR and can be accelerated by reducing the number of TR’s. Our proposed trajectory takes advantage of the 4D-encoding space by spreading incoherent aliasing across spatial and spectral dimensions.