Yikang Li1,2,3, Zhan li Hu1,2,3, Sen Jia1,2,3, Wenjing Xu4, Zongyang Li1,2,3, Hairong Zheng1,2,3, Xin Liu1,2,3, and Na Zhang1,2,3
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 3CAS key laboratory of health informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 4Faculty of Information Technology, Beijing University of Technology, Beijng, China
1Paul C. Lauterbur Research Center for Biomedical Imaging, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 2Key Laboratory for Magnetic Resonance and Multimodality Imaging of Guangdong Province, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 3CAS key laboratory of health informatics, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen, China, 4Faculty of Information Technology, Beijing University of Technology, Beijng, China
We proposed a new method that omits the Fourier transformations and directly makes segmentations on K space data.
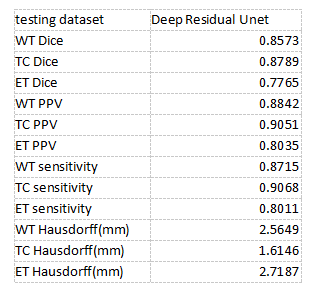
Table 1. The testing dataset results. Mean Dice, PPV, sensitivity, and Hausdorff are shown in the table. EN is enhancing tumor core. WT is the whole tumor part. And the TC is tumor core.
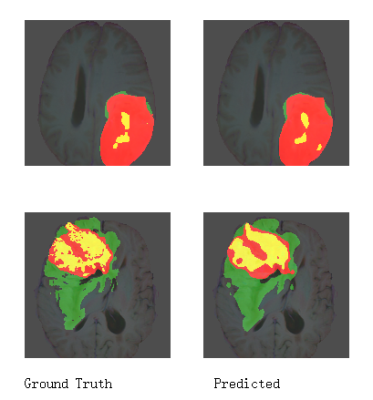
Figure 2. A typical segmentation example with ground truth and predicted labels on 2D MRI slices. The whole tumor (WT) part contains all visible labels (all green, red and yellow labels), the tumor core (TC) part contains red and yellow labels, and the enhancing tumor core (ET) class is shown in yellow.