Chantal Tax1,2, Hugo Larochelle3, Joao P. De Almeida Martins4, Jana Hutter5, Derek K. Jones2, Maxime Chamberland2, and Maxime Descoteaux6
1Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 2CUBRIC, Cardiff University, Cardiff, United Kingdom, 3Google Brain, Montreal, QC, Canada, 4Department of Radiology and Nuclear Medicine, St. Olav's University Hospital, Trondheim, Norway, 5Centre for Medical Engineering, King's College London, London, United Kingdom, 6SCIL, University of Sherbrooke, Sherbrooke, QC, Canada
1Image Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 2CUBRIC, Cardiff University, Cardiff, United Kingdom, 3Google Brain, Montreal, QC, Canada, 4Department of Radiology and Nuclear Medicine, St. Olav's University Hospital, Trondheim, Norway, 5Centre for Medical Engineering, King's College London, London, United Kingdom, 6SCIL, University of Sherbrooke, Sherbrooke, QC, Canada
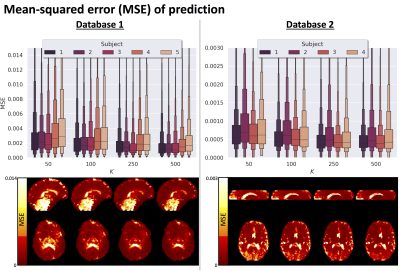
Multi-contrast MRI provides a comprehensive picture of tissue microstructure, but the high dimensionality of the parameter space increases scan time. In this work, we present a data-driven approach to multi-contrast MRI experiment design using unsupervised machine learning.
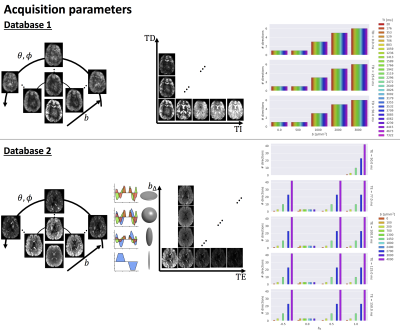
Sampling for both databases, varying inversion time (TI), delay time (TD), (θ,φ), b-value, and b∆ parameterising the ”shape” of the diffusion-encoding b-tensor16 (represented by the ellipsoids, with corresponding encoding waveforms along each axis). Left: images are scaled per volume except in the TD/TE dimension. Right: number of directions for each parameter setting.