Nina Elina Hänninen1,2, Mikko Johannes Nissi1,2, Matti Hanni1,3,4, Olli Gröhn5, Miika Tapio Nieminen1,3,4, and Timo Liimatainen1,4
1Research Unit of Medical Imaging, Physics and Technology, University of Oulu, Oulu, Finland, 2Department of Applied Physics, University of Eastern Finland, Kuopio, Finland, 3Medical Research Center, University of Oulu and Oulu University Hospital, Oulu, Finland, 4Department of Diagnostic Radiology, Oulu University Hospital, Oulu, Finland, 5A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland, Kuopio, Finland
1Research Unit of Medical Imaging, Physics and Technology, University of Oulu, Oulu, Finland, 2Department of Applied Physics, University of Eastern Finland, Kuopio, Finland, 3Medical Research Center, University of Oulu and Oulu University Hospital, Oulu, Finland, 4Department of Diagnostic Radiology, Oulu University Hospital, Oulu, Finland, 5A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland, Kuopio, Finland
Cartilage and
tendon represented highest relaxation anisotropy in comparison to brain, spinal
cord, kidney and cardiac muscle tissue. T2 presented highest anisotropy, while
rotating frame relaxations exhibited less anisotropy, and T1 showed no
anisotropy.
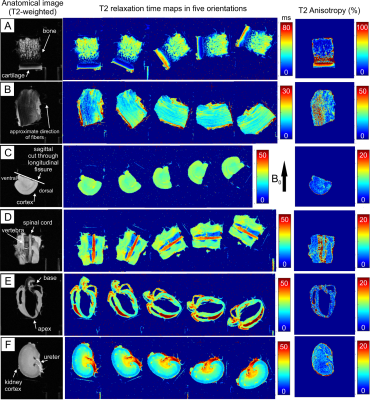
Figure
1. T2-weighted images (left), T2 maps in 5 different orientations with respect
to B0 (middle), and calculated T2 relaxation anisotropy maps (right) for
representative tissue samples: A) Articular cartilage (bovine patella); B) Tendon
(bovine ACL); C) Brain tissue (mouse, right hemisphere, coronal view); D) Spinal
cord and muscle (mouse, sagittal view); E) Cardiac muscle (mouse, coronal view);
F) Kidney (mouse, coronal view). The color scales are adjusted separately for
each sample.
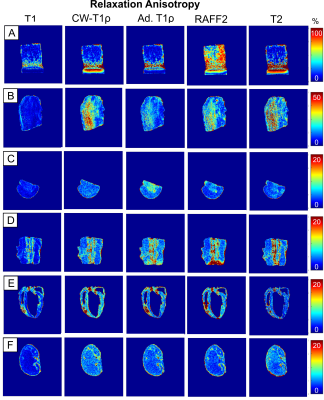
Figure 2. Relaxation anisotropy maps of
different parameters, T1, CW-T1ρ (500 Hz), adiabatic T1ρ, RAFF2, and T2 in
representative tissue samples: A) Articular cartilage (bovine patella); B)
Tendon (bovine ACL); C) Brain tissue (mouse, right hemisphere, coronal view); D)
Spinal cord and muscle (mouse, sagittal view); E) Cardiac muscle (mouse,
coronal view); F) Kidney (mouse, coronal view). The color scales are adjusted separately
for each sample.
