Jan Kufer1, Christine Preibisch1,2, Samira Epp1, Jens Goettler1,3, Kilian Weiss4, Mikkel Bo Hansen5, Claus Zimmer1, Kim Mouridsen5, Fahmeed Hyder3, and Stephan Kaczmarz1,3
1Department of Neuroradiology, School of Medicine, Technical University of Munich (TUM), Munich, Germany, 2Department of Neurology, School of Medicine, Technical University of Munich (TUM), Munich, Germany, 3Department of Radiology & Biomedical Imaging (MRRC), Yale University, New Haven, CT, United States, 4Philips Healthcare, Hamburg, Germany, 5Department of Clinical Medicine, Aarhus University, Aarhus, Denmark
1Department of Neuroradiology, School of Medicine, Technical University of Munich (TUM), Munich, Germany, 2Department of Neurology, School of Medicine, Technical University of Munich (TUM), Munich, Germany, 3Department of Radiology & Biomedical Imaging (MRRC), Yale University, New Haven, CT, United States, 4Philips Healthcare, Hamburg, Germany, 5Department of Clinical Medicine, Aarhus University, Aarhus, Denmark
Comparison
of two MRI-based cerebral oxygenation biomarkers OEF from mqBOLD and OEC modelled
from DSC showed good agreement with PET reference data in cortical gray matter in
young healthy volunteers. Some variation between MRI-based OEF and OEC was attributed to different underlying models.
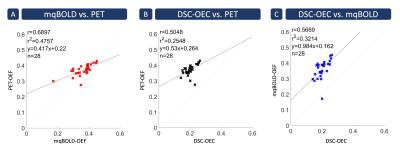
Figure 3: Spatial
correlation across 28 gray matter VOIs. Each cross indicates the mean parameter value in a specific Brodmann
area averaged across subjects. Strongest spatial correlation was found between
PET- and mqBOLD-OEF (A) with r=0.69 (p<0.05). Correlation between PET-OEF
and DSC-OEC (B) was also strong, but slightly weaker with r=0.50 (p<0.05). Similarly,
good correlation was also found between both MRI techniques (C) with r=0.57 (p<0.05).
Note that MRI and PET data were acquired in similar cohorts, but different
subjects.
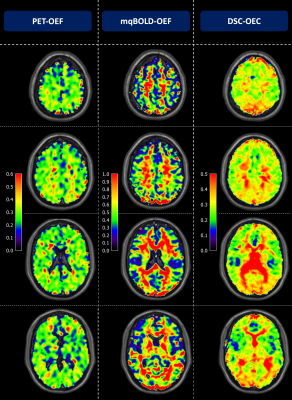
Figure 2: Exemplary PET-OEF, MRI-OEF and MRI-OEC data in
four different slices. PET-OEF
(left column) is quite homogenous across the entire brain, without contrast
between gray (GM) vs. white matter (WM). MRI-based OEF (central column) and OEC
(right column) show some similarity with PET. However, both parameter maps show
an artifactual GM/WM contrast with elevated WM values. While OEF values in GM
seem similar between PET and mqBOLD, OEC values based on the CFIN-model are
generally lower.
